This is the multi-page printable view of this section. Click here to print.
Spectroscopy
1 - Spectroscopy with lcmodel
This tutorial was created by Steffen Bollmann.
Github: @stebo85 Web: mri.sbollmann.net Twitter: @sbollmann_MRI
Getting Setup with Neurodesk
For more information on getting set up with a Neurodesk environment, see hereOpen lcmodel from the menu: Applications -> Spectroscopy -> lcmodel -> lcmodel 6.3
run
setup_lcmodel.sh
then run
lcmgui
We packed example data into the container (https://zenodo.org/record/3904443/) and we will use this to show a basic analysis.
The example data comes in the Varian fid format, so click on Varian:
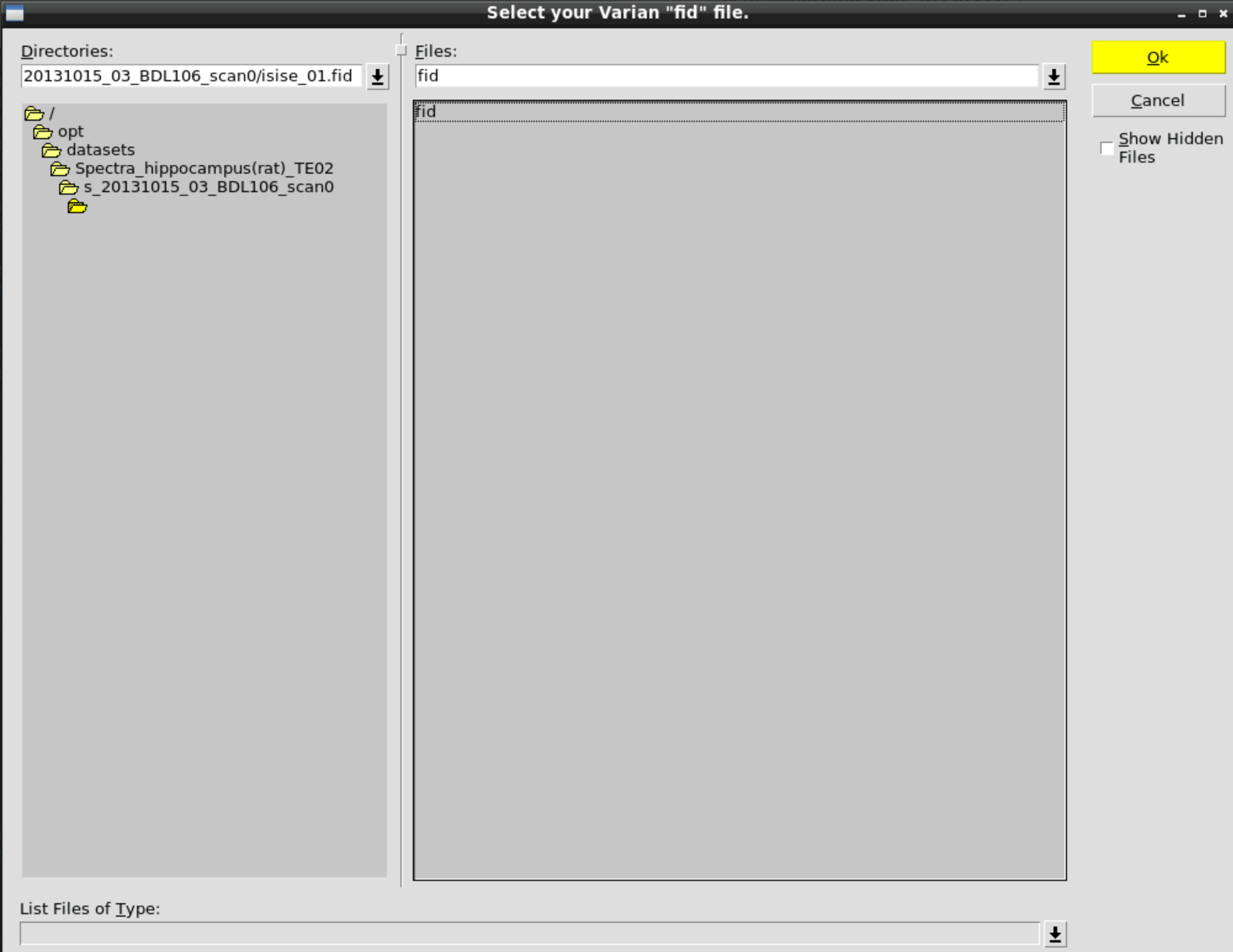
and then select the fid data in: /opt/datasets/Spectra_hippocampus(rat)_TE02/s_20131015_03_BDL106_scan0/isise_01.fid
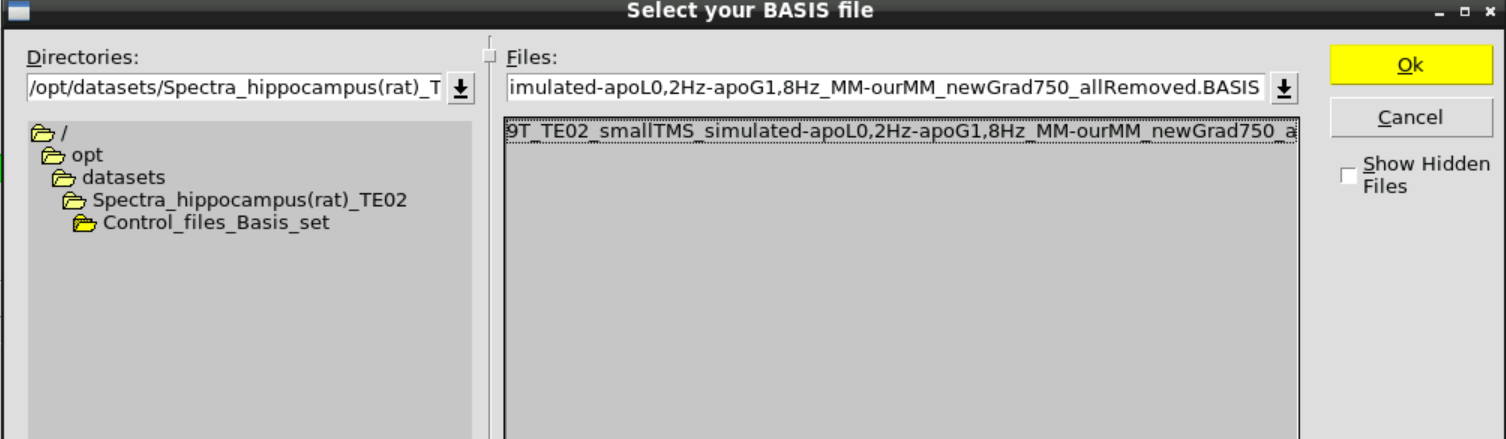
Then Change BASIS and select the appropriate basis set in /opt/datasets/Spectra_hippocampus(rat)_TE02/Control_files_Basis_set
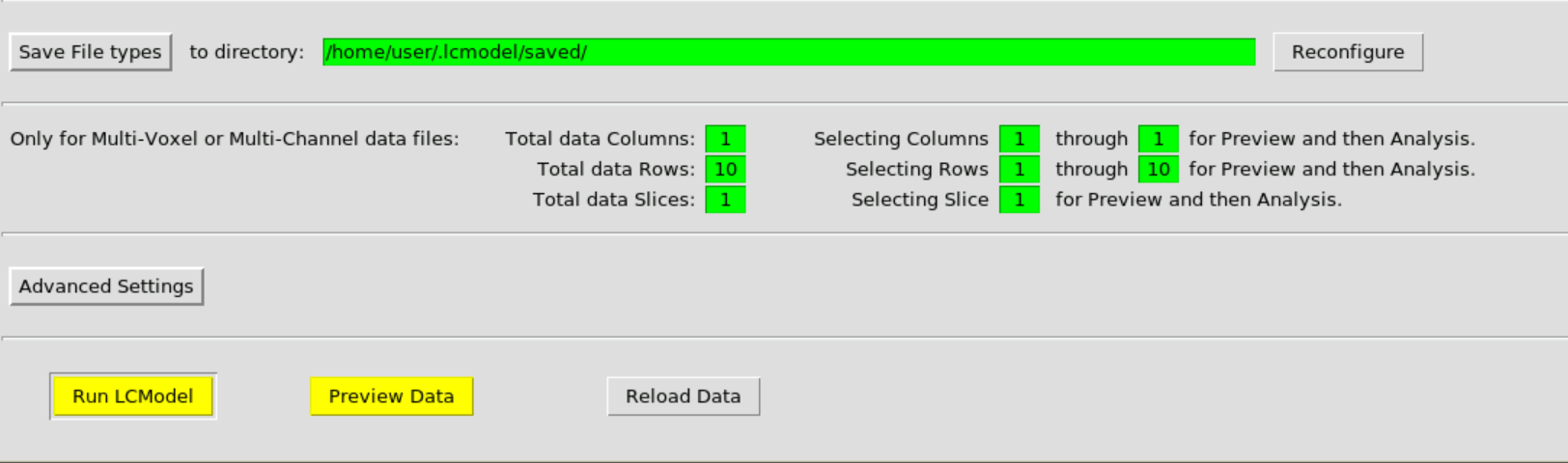
Then hit Run LCModel:
and confirm:
then wait a couple of minutes until the analyzed spectra appear - by closing the window you can go through the results:
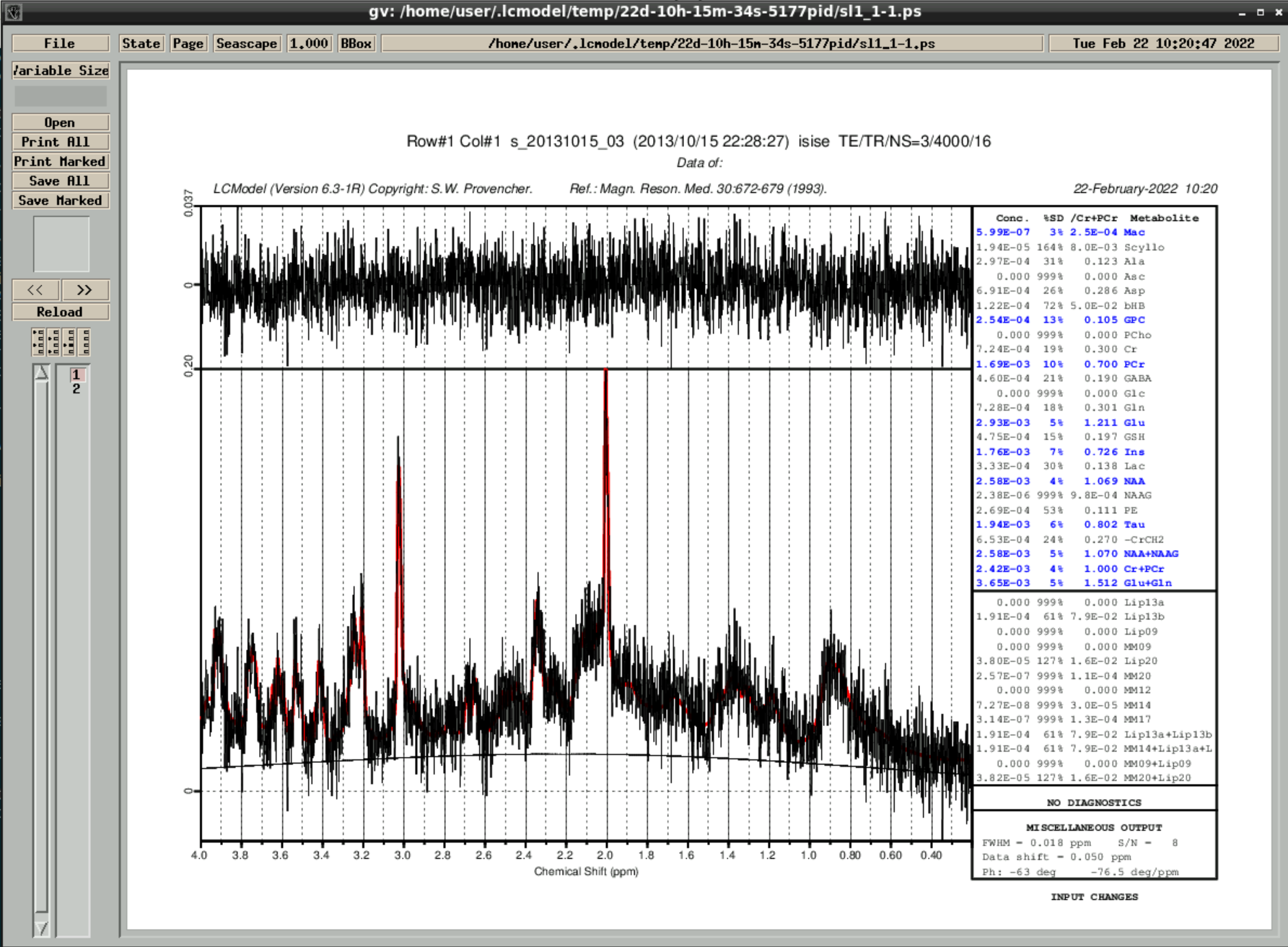
the results are also saved in ~/.lcmodel/saved/
2 - Spectroscopy pipeline
This tutorial was created by Korbinian Eckstein.
Github: @korbinian90
Getting Setup with Neurodesk
For more information on getting set up with a Neurodesk environment, see hereProcessing MRSI
ml mrsiproc/0.1.0
bash Part1_ProcessMRSI.sh [ARGUMENTS]
bash Part2_EvaluateMRSI.sh [ARGUMENTS]
Starting Neurodesk
After starting Neurodesk, a JupyterLab instance should open. You can either work from here or open a desktop environment by clicking Neurodesktop under Notebooks. This tutorial uses the desktop.
Running MRSI reconstruction
Open vscode and create and open a new folder under neurodesktop-storage
Info
The processing can either start from reconstructed DICOM files or exported dat files. A reconstruction of a dat files might look like this:
Part1_ProcessMRSI.sh \
-c /neurodesktop-storage/data/meas_test.dat \
-t /neurodesktop-storage/data/T1/ \
-a /neurodesktop-storage/data/INV1/ \
-b fid_1.300000ms.basis \
-o /neurodesktop-storage/mrsi_proc/ProcessResults/test_files \
-j LCModel_Control_Template.m \
-m "dreid"
The fid_1.300000ms.basis and LCModel_Control_Template.m files are included
Here, we create a new bash file with the default settings and execute it. Copy the contents of this template file to run_mrsi_part1.sh and replace the -c, -t and -a arguments with your own data and set the output path with -o.
#!/bin/bash
# There are two script to process the mrsi data
# First run this script, then run the part 2 script
ml mrsiproc/0.1.0
## The required files are:
# - DAT file / DICOM files
# - T1 (DICOM or .mnc)
## Create own mask (not fully supported yet)
# hd-bet -i /neurodesktop-storage/mrsi_proc/test_files/MP2RAGE/t1.nii.gz -o /neurodesktop-storage/mrsi_proc/test_files/mask.nii.gz -device cpu -mode fast -tta 0
# nii2mnc /neurodesktop-storage/mrsi_proc/test_files/mask.nii.gz /neurodesktop-storage/mrsi_proc/test_files/mask.mnc
Part1_ProcessMRSI.sh \
-c /neurodesktop-storage/data/meas_test.dat \
-t /neurodesktop-storage/data/T1/ \
-a /neurodesktop-storage/data/INV1/ \
-b fid_1.300000ms.basis \
-o /neurodesktop-storage/mrsi_proc/ProcessResults/test_files \
-j LCModel_Control_Template.m \
-m "dreid" \
-L "L2,0.2"
## HINT
# To change the number of threads used by LCModel, change the number in the LCModel_Control_Template.m file (default 8)
## WARNING -s
# The Julia based recontstruction is still experimental! The algorithm is different, and the results are not expected to be identical. It should be identical to the DICOM output from the ICE version.
## WARNING -m
# If a mask file is given, it must be in *.mnc format
# The word "bet" may not be part of the filename of the mask given to the -m flag
# For a 3D mask, it is suggested to use -m "dreid" instead of -m "bet"
## WARNING -t
# The DICOM folder for giving the T1-weighted image must not contain any other files than *.IMA
# The t1-weighted file can be given as *.mnc file as well
## The OPTIONS are
# Usage: %s
# mandatory:
# -c [csi file] Format: DAT, DICOM, or .mat. If a .mat file is passed over, it is expected that everything is already performed like coil combination etc.
# You can pass over several files of the same type by \'-c \"[csi_path1] [csi_path2] ...\"\'. These files get individually processed and averaged
# at the end.
# -b [basis files] Format: .BASIS. Used for LCM fitting (for FID)
# -B [basis files] Format: .BASIS. Used for LCM fitting (spin echo: for fidesi = fid + echo)
# -o [output directory]
# optional:
# -i [image NORMAL] Format: DAT or DICOM. The FoV must match that of the CSI file. Used for our coil combination and for creating mask (if no T1 is inputted)
# -f [image FLIP] Format: DAT or DICOM. Imaging file FLIP (FOV rotated about -180 deg). Used for correcting gradient delays.
# -v [VC image] Format: DAT or DICOM. Image of volume or body coil file. Used for sensmap method or for creating mask.
# -t [T1 images] Format: DICOM. Folder of 3d T1-weighted acquisition containing DICOM files. Used for creating mask and for visual purposes. If minc file is given instead of folder, it is treated as the magnitude file.
# -a [T1 AntiNoise images] Format: DICOM. Folder of 3d T1-weighted acquisition containing DICOM files. Used for pre-masking the T1w image to get rid of the noise in air-areas.
# -w [Water Reference] Format: DAT or DICOM. LCModel 'Do Water Scaling' or separate water quantification (Water maps are created). The same scan as -c [csi file], but without water suppression.
# -m [mask] Defines how to create the mask. Options: -m \"bet\", \"thresh\", \"voi\", \"[Path_to_usermade_mask]\". If not set --> no mask used.
# -h [100] Hamming filter CSI data.
# -r [InPlaneCaipPattern_And_VD_Radius] The InPlaneCaipPattern and the VD_Radius as used in ParallelImagingSimReco.m. Example: \"InPlaneCaipPattern = [0 0 0; 0 0 0; 0 0 1]; VD_Radius = 2;\".
# -R [SliceAliasingPattern]
# -g [noisedecorr_path] If this option is used the csi data gets noise decorrelated using noise from passed-over noise file, or if -g \" is given, by noise from the end of the FIDs at the border of the FoV or from the PRESCAN, if available.
# -F [Nothing] If this option is set, the spectra are corrected for the first order phase caused by an acquisition delay of the FID-sequences. You must provide a basis set with an appropriate acquisition delay. DONT USE WITH SPIN ECHO SEQUENCES.
# -u [Nothing] If a phantom was measured. Different settings used for fitting (e.g. some metabolites are omitted)
# -I [\"nextpow2\" Or Vector]If nextpow2: Perform zerofilling to the next power of 2 in ROW and COL dimensions (e.g. from 42x42 to 64x64). If vector (e.g. [16 16 1]): Spatially Interpolate to this size.
# -A [\"\" Or Path] Perform frequency alignment. If a mnc file is given, use these as B0-map, otherwise shift according to water peak of center voxel.
# -l [Nothing] If this option is set, LCModel is not started, everything else is done normally. Useful for only computing the SNR.
# -j [LCM_ControlFile] ControlFile telling LCModel how to process the data. for FID
# -J [LCM_ControlFile] ControlFile telling LCModel how to process the data. for ECHO
# -X [XPACE MOTION LOG] XPACE MOTION LOG
# otherwise standard values are assumed. A template file is provided in this package.
# -e [LineBroadeningInHz] Apply an exponential filter to the spectra [Hz].
# -s [threads] [mmap] Use the Julia reconstruction version (less RAM usage, different reconstruction algorithm). [threads=auto] can be auto or a number. [mmap=false] can be \"true\", \"false\" or a path.
# -d [Nothing] Use the deprecated, old dat file format (before sequence merging, 06/2023)
Then run the script with
bash run_mrsi_part1.sh
This can take several hours for reconstruction and LCModel processing.
Part 2
Continue with the same process for part 2
Template file for part 2
#!/bin/bash
ml mrsiproc/0.1.0
Part2_EvaluateMRSI.sh \
-o neurodesktop-storage/mrsi_proc/ProcessResults/test_files \
-S "Glu,Gln,Raw-abs-csi" \
-N Nifti \
-R \
-b
## The OPTIONS are
# Usage: %s
# mandatory:
# -o [output directory]
# optional:
# -d [print_indiv..._flag] If this option is set, the SNR gets computed by our own program. If the
# print_individual_spectra_flag=1 (by using option -d 1) all spectra for
# computing the SNR are printed.
# -b [segmentation_matrix_size]
# If segmentation to GM, WM and CSF should be performed.
# -s [CRLB_treshold_value] user can set the treshold value for CRLB in the metabolic maps
# -n [SNR_treshold_value] If this option is set (user set the value of SNR threshold after the flag),
# SNR binary mask is computed either for LC model SNR or (if the -d flag is set) for
# custom SNR computation method and LCmodel
# -f [FWHM_treshold_value] If this option is set (user set the value of FWHM threshold after the flag),
# FWHM binary mask is computed from LCmodel results
# -a [Control file] Compute SNR with home-brewed script.
# -l [local_folder] Perform some of the file-heavy tasks in \$local_folder/tmpx (x=1,2,3,...) instead of on \$out_dir
# directly. This is faster, and if \$out_dir is mounted via nfs4, writing directly to \$out_dir
# can lead to timeouts and terrible zombie processes.
# -k [spectra_stack_range] If this option is set, the .Coord files from LCmodel are used to create stacks of spectra
# to visualize the fit in corresponding voxels (results stored in form of .eps)
# user can set the starting point of range for stack of spectra for display purposes (in ppm)
# user can set the ending point of range for stack of spectra for display purposes (in ppm)
# Options: \"['fullrange']\", \"[ppm_start; ppm_end]\"
# -r [non_lin_reg_type] If this option is set, the non-linear registration is computed using minctools, Options: -r \"MNI305\", \"MNI152\"
# -w [compute_reg_only_flag] If this option is set, only the non-linear registration is computed.
# -q [compute_seg_only_flag] If this option is set, only the segmentation is computed.
# -N [\"Nifti\" or \"Both\"] Only create nifti-files. If \"Both\" option is used, create minc and nifti. If -N option is not used, creat only mnc files.
# -S [SpectralMap_flag] Create a nifti-file with a map of the LCModel-spectra and -fits to view with freeview as timeseries.\nNeeds freesurfer-linux-centos7_x86_64-dev-20181113-8abd50c, and MATLAB version > R2017b. Can only be used with -N option.
# -u [UpsampledMaps_flag] Create upsampled maps by zero-filling (in future more sophisticated methods might be implemented).
# -R [RatioMaps_flag] Create Ratio maps.