AFNI Preprocessing#
Author: Monika Doerig
Date: 1 May 2025
Citation and Resources:#
Tools included in this workflow#
AFNI
Cox RW (1996). AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29(3):162-173. doi:10.1006/cbmr.1996.0014
RW Cox, JS Hyde (1997). Software tools for analysis and visualization of FMRI Data. NMR in Biomedicine, 10: 171-178. https://pubmed.ncbi.nlm.nih.gov/9430344/ 15.1.2. Methods: General func
Educational Resources#
Andy’s Brain Book:
This AFNI example is based on the AFNI Tutorial: Commands and Preprocessing from Andy’s Brain Book (Jahn, 2022. doi:10.5281/zenodo.5879293)
Dataset#
Flanker Dataset from OpenNeuro:
Kelly AMC and Uddin LQ and Biswal BB and Castellanos FX and Milham MP (2018). Flanker task (event-related). OpenNeuro Dataset ds000102. [Dataset] doi: null
Kelly AM, Uddin LQ, Biswal BB, Castellanos FX, Milham MP. Competition between functional brain networks mediates behavioral variability. Neuroimage. 2008 Jan 1;39(1):527-37. doi: 10.1016/j.neuroimage.2007.08.008. Epub 2007 Aug 23. PMID: 17919929.
Mennes, M., Kelly, C., Zuo, X.N., Di Martino, A., Biswal, B.B., Castellanos, F.X., Milham, M.P. (2010). Inter-individual differences in resting-state functional connectivity predict task-induced BOLD activity. Neuroimage, 50(4):1690-701. doi: 10.1016/j.neuroimage.2010.01.002. Epub 2010 Jan 15. Erratum in: Neuroimage. 2011 Mar 1;55(1):434
Mennes, M., Zuo, X.N., Kelly, C., Di Martino, A., Zang, Y.F., Biswal, B., Castellanos, F.X., Milham, M.P. (2011). Linking inter-individual differences in neural activation and behavior to intrinsic brain dynamics. Neuroimage, 54(4):2950-9. doi: 10.1016/j.neuroimage.2010.10.046
Load AFNI#
import module
await module.load('afni/21.2.00')
await module.list()
['afni/21.2.00']
Import Python Modules#
%%capture
!pip install nibabel numpy nilearn
import os
import nibabel as nib
import numpy as np
import matplotlib.pyplot as plt
from ipyniivue import NiiVue
from IPython.display import display
from ipywidgets import VBox, Dropdown
import ipywidgets as widgets
from IPython.display import Image
from nilearn.image import index_img
Data preparation#
PATTERN = "sub-08"
!datalad install https://github.com/OpenNeuroDatasets/ds000102.git
!cd ds000102 && datalad get $PATTERN
Cloning: 0%| | 0.00/2.00 [00:00<?, ? candidates/s]
Enumerating: 0.00 Objects [00:00, ? Objects/s]
Counting: 0%| | 0.00/27.0 [00:00<?, ? Objects/s]
Compressing: 0%| | 0.00/23.0 [00:00<?, ? Objects/s]
Receiving: 0%| | 0.00/2.15k [00:00<?, ? Objects/s]
Resolving: 0%| | 0.00/537 [00:00<?, ? Deltas/s]
[INFO ] scanning for unlocked files (this may take some time)
[INFO ] Remote origin not usable by git-annex; setting annex-ignore
[INFO ] access to 1 dataset sibling s3-PRIVATE not auto-enabled, enable with:
| datalad siblings -d "/home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/ds000102" enable -s s3-PRIVATE
install(ok): /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/ds000102 (dataset)
Total: 0%| | 0.00/67.8M [00:00<?, ? Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 0%| | 0.00/10.6M [00:00<?, ? Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 1%| | 138k/10.6M [00:00<00:17, 593k Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 3%|▏ | 364k/10.6M [00:00<00:10, 1.02M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 8%|▎ | 869k/10.6M [00:00<00:04, 2.27M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 16%|▍ | 1.74M/10.6M [00:00<00:02, 4.25M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 32%|▉ | 3.41M/10.6M [00:00<00:00, 8.02M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 58%|█▋ | 6.11M/10.6M [00:00<00:00, 13.5M Bytes/s]
Get sub-08/a .. 8_T1w.nii.gz: 81%|██▍| 8.50M/10.6M [00:00<00:00, 16.6M Bytes/s]
Total: 16%|████ | 10.6M/67.8M [00:01<00:08, 6.54M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 0%| | 0.00/28.6M [00:00<?, ? Bytes/s]
Get sub-08/f .. _bold.nii.gz: 5%|▏ | 1.42M/28.6M [00:00<00:02, 11.3M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 13%|▍ | 3.71M/28.6M [00:00<00:02, 11.3M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 21%|▋ | 6.10M/28.6M [00:00<00:01, 15.2M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 32%|▉ | 9.27M/28.6M [00:00<00:01, 15.5M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 41%|█▏ | 11.8M/28.6M [00:00<00:00, 17.9M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 50%|█▍ | 14.3M/28.6M [00:00<00:00, 19.7M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 58%|█▋ | 16.6M/28.6M [00:00<00:00, 20.8M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 67%|██ | 19.2M/28.6M [00:01<00:00, 22.2M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 75%|██▎| 21.5M/28.6M [00:01<00:00, 22.4M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 92%|██▋| 26.2M/28.6M [00:01<00:00, 22.9M Bytes/s]
Total: 58%|███████████████ | 39.2M/67.8M [00:03<00:02, 11.6M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 0%| | 0.00/28.6M [00:00<?, ? Bytes/s]
Get sub-08/f .. _bold.nii.gz: 5%|▏ | 1.45M/28.6M [00:00<00:02, 9.44M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 15%|▍ | 4.42M/28.6M [00:00<00:02, 11.9M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 24%|▋ | 6.87M/28.6M [00:00<00:01, 15.6M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 32%|▉ | 9.20M/28.6M [00:00<00:01, 17.9M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 47%|█▍ | 13.4M/28.6M [00:00<00:00, 19.5M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 55%|█▋ | 15.7M/28.6M [00:00<00:00, 20.1M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 63%|█▉ | 18.2M/28.6M [00:00<00:00, 21.3M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 79%|██▎| 22.5M/28.6M [00:01<00:00, 21.5M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 87%|██▌| 25.0M/28.6M [00:01<00:00, 22.3M Bytes/s]
Get sub-08/f .. _bold.nii.gz: 96%|██▉| 27.6M/28.6M [00:01<00:00, 23.0M Bytes/s]
get(ok): sub-08/anat/sub-08_T1w.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-08/func/sub-08_task-flanker_run-1_bold.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-08/func/sub-08_task-flanker_run-2_bold.nii.gz (file) [from s3-PUBLIC...]
get(ok): sub-08 (directory)
action summary:
get (ok: 4)
The data is structured in BIDS format:
!tree -L 4 ds000102
ds000102
├── CHANGES
├── README
├── T1w.json
├── dataset_description.json
├── derivatives
│ └── mriqc
│ ├── aMRIQC.csv -> ../../.git/annex/objects/Q4/jv/MD5E-s14180--3addf0456b803b7c5ec5147481ecdd62.csv/MD5E-s14180--3addf0456b803b7c5ec5147481ecdd62.csv
│ ├── anatomical_group.pdf -> ../../.git/annex/objects/6m/q9/MD5E-s98927--d11151f65ae061833e7fd4373adfec3f.pdf/MD5E-s98927--d11151f65ae061833e7fd4373adfec3f.pdf
│ ├── anatomical_sub-01.pdf -> ../../.git/annex/objects/K3/7x/MD5E-s2747349--5d40f2a54fb4194ac4a79f0295ff51c0.pdf/MD5E-s2747349--5d40f2a54fb4194ac4a79f0295ff51c0.pdf
│ ├── anatomical_sub-02.pdf -> ../../.git/annex/objects/Kx/Kv/MD5E-s2803965--56f6b768362bd9b7f0ef501b8cb6dde6.pdf/MD5E-s2803965--56f6b768362bd9b7f0ef501b8cb6dde6.pdf
│ ├── anatomical_sub-03.pdf -> ../../.git/annex/objects/kx/g5/MD5E-s2809843--e90d7a4859ed4be986b55e23f93ca89d.pdf/MD5E-s2809843--e90d7a4859ed4be986b55e23f93ca89d.pdf
│ ├── anatomical_sub-04.pdf -> ../../.git/annex/objects/JK/Zm/MD5E-s2846770--4869146771178dbb01ac79b95b35a8a1.pdf/MD5E-s2846770--4869146771178dbb01ac79b95b35a8a1.pdf
│ ├── anatomical_sub-05.pdf -> ../../.git/annex/objects/zm/FG/MD5E-s2824086--fda634d34556c83005a5eb2ca8c498dd.pdf/MD5E-s2824086--fda634d34556c83005a5eb2ca8c498dd.pdf
│ ├── anatomical_sub-06.pdf -> ../../.git/annex/objects/92/q0/MD5E-s2798058--fdddf0aff1eca8f61ed7c8b04ada9735.pdf/MD5E-s2798058--fdddf0aff1eca8f61ed7c8b04ada9735.pdf
│ ├── anatomical_sub-07.pdf -> ../../.git/annex/objects/39/3K/MD5E-s2795270--29ce2e2352596df940e5f3fae45b5a38.pdf/MD5E-s2795270--29ce2e2352596df940e5f3fae45b5a38.pdf
│ ├── anatomical_sub-08.pdf -> ../../.git/annex/objects/Fx/F4/MD5E-s2727492--b55dad8ffe22fc035110ecf4119d2960.pdf/MD5E-s2727492--b55dad8ffe22fc035110ecf4119d2960.pdf
│ ├── anatomical_sub-09.pdf -> ../../.git/annex/objects/4M/pz/MD5E-s2887144--28ea830af2a4d741147d18ea9c7fda84.pdf/MD5E-s2887144--28ea830af2a4d741147d18ea9c7fda84.pdf
│ ├── anatomical_sub-10.pdf -> ../../.git/annex/objects/0z/Vw/MD5E-s2874045--6542a57a9fc58f97f2a03c2384663c62.pdf/MD5E-s2874045--6542a57a9fc58f97f2a03c2384663c62.pdf
│ ├── anatomical_sub-11.pdf -> ../../.git/annex/objects/wm/76/MD5E-s2781221--1071b83e3c1b4532879521c37c3329da.pdf/MD5E-s2781221--1071b83e3c1b4532879521c37c3329da.pdf
│ ├── anatomical_sub-12.pdf -> ../../.git/annex/objects/GF/19/MD5E-s2817233--bfd24ca3274fa5efd654e2afd927f9ef.pdf/MD5E-s2817233--bfd24ca3274fa5efd654e2afd927f9ef.pdf
│ ├── anatomical_sub-13.pdf -> ../../.git/annex/objects/9Q/X2/MD5E-s2796088--ed299ab7e1662cb03aa01299eed2602b.pdf/MD5E-s2796088--ed299ab7e1662cb03aa01299eed2602b.pdf
│ ├── anatomical_sub-14.pdf -> ../../.git/annex/objects/Wg/55/MD5E-s2558074--cadc9bd81856dcd02677de84e7e6ca90.pdf/MD5E-s2558074--cadc9bd81856dcd02677de84e7e6ca90.pdf
│ ├── anatomical_sub-15.pdf -> ../../.git/annex/objects/43/Q3/MD5E-s2847293--0c678a4b309d055ad9ba4ba25b77351b.pdf/MD5E-s2847293--0c678a4b309d055ad9ba4ba25b77351b.pdf
│ ├── anatomical_sub-16.pdf -> ../../.git/annex/objects/xq/qj/MD5E-s2890454--87c62253c1711f30d53c41b3ac38dc66.pdf/MD5E-s2890454--87c62253c1711f30d53c41b3ac38dc66.pdf
│ ├── anatomical_sub-17.pdf -> ../../.git/annex/objects/zK/M1/MD5E-s2825765--0a91015e22836a3076641b963e1ccfc6.pdf/MD5E-s2825765--0a91015e22836a3076641b963e1ccfc6.pdf
│ ├── anatomical_sub-18.pdf -> ../../.git/annex/objects/w2/Vk/MD5E-s2821624--1d9a3f0b21ce1f9a3b490d44d36f1f11.pdf/MD5E-s2821624--1d9a3f0b21ce1f9a3b490d44d36f1f11.pdf
│ ├── anatomical_sub-19.pdf -> ../../.git/annex/objects/J2/Jq/MD5E-s2453814--533411f3353cb3fa0264485e81f3fcf6.pdf/MD5E-s2453814--533411f3353cb3fa0264485e81f3fcf6.pdf
│ ├── anatomical_sub-20.pdf -> ../../.git/annex/objects/MF/9X/MD5E-s2881144--176c560778c55db87e8468b3246d373c.pdf/MD5E-s2881144--176c560778c55db87e8468b3246d373c.pdf
│ ├── anatomical_sub-21.pdf -> ../../.git/annex/objects/XQ/p1/MD5E-s2330589--b7546dfe5fb43a974cd23111b860c493.pdf/MD5E-s2330589--b7546dfe5fb43a974cd23111b860c493.pdf
│ ├── anatomical_sub-22.pdf -> ../../.git/annex/objects/Fx/k8/MD5E-s2505165--55f0661ad209b742c517cc5b5469436a.pdf/MD5E-s2505165--55f0661ad209b742c517cc5b5469436a.pdf
│ ├── anatomical_sub-23.pdf -> ../../.git/annex/objects/qj/8K/MD5E-s2784018--7e8697a7d4601547a899a27af132166d.pdf/MD5E-s2784018--7e8697a7d4601547a899a27af132166d.pdf
│ ├── anatomical_sub-24.pdf -> ../../.git/annex/objects/G8/Kw/MD5E-s2828817--e86be931adef2a7b0297d557d827d629.pdf/MD5E-s2828817--e86be931adef2a7b0297d557d827d629.pdf
│ ├── anatomical_sub-25.pdf -> ../../.git/annex/objects/XG/kg/MD5E-s2447908--3d392b9d27929dc4146d2b47be16e8dc.pdf/MD5E-s2447908--3d392b9d27929dc4146d2b47be16e8dc.pdf
│ ├── anatomical_sub-26.pdf -> ../../.git/annex/objects/8P/42/MD5E-s2850007--6d2f87a305b30d5704aaf4be9b8ff1e6.pdf/MD5E-s2850007--6d2f87a305b30d5704aaf4be9b8ff1e6.pdf
│ ├── fMRIQC.csv -> ../../.git/annex/objects/2Z/Ff/MD5E-s21038--cbe73db3db1beb0a1977583cff2a724b.csv/MD5E-s21038--cbe73db3db1beb0a1977583cff2a724b.csv
│ ├── functional_group.pdf -> ../../.git/annex/objects/Kq/xg/MD5E-s90712--7058c3db328fecb86303bc27a9ef0110.pdf/MD5E-s90712--7058c3db328fecb86303bc27a9ef0110.pdf
│ ├── functional_sub-01.pdf -> ../../.git/annex/objects/k2/vQ/MD5E-s1157925--e055f942b72b9aabad7a5e3d7b25b201.pdf/MD5E-s1157925--e055f942b72b9aabad7a5e3d7b25b201.pdf
│ ├── functional_sub-02.pdf -> ../../.git/annex/objects/X3/X6/MD5E-s1235840--cb32b7f8f1274af250b4f0fc15dacecb.pdf/MD5E-s1235840--cb32b7f8f1274af250b4f0fc15dacecb.pdf
│ ├── functional_sub-03.pdf -> ../../.git/annex/objects/Vp/0x/MD5E-s1228507--73ab1cc4cb27712892fcb10a0853ba7c.pdf/MD5E-s1228507--73ab1cc4cb27712892fcb10a0853ba7c.pdf
│ ├── functional_sub-04.pdf -> ../../.git/annex/objects/xk/jz/MD5E-s1252659--8ae6d1b02767c1ddb72dd7e6afefe696.pdf/MD5E-s1252659--8ae6d1b02767c1ddb72dd7e6afefe696.pdf
│ ├── functional_sub-05.pdf -> ../../.git/annex/objects/Zm/VJ/MD5E-s1258815--dd043691d548a501dd63d1aaf420e43c.pdf/MD5E-s1258815--dd043691d548a501dd63d1aaf420e43c.pdf
│ ├── functional_sub-06.pdf -> ../../.git/annex/objects/M5/gq/MD5E-s1247345--9c06bc69792b812ab8deffb01c6656c2.pdf/MD5E-s1247345--9c06bc69792b812ab8deffb01c6656c2.pdf
│ ├── functional_sub-07.pdf -> ../../.git/annex/objects/25/35/MD5E-s1229731--99cc64e99df0025ccb0341cd0dcf688b.pdf/MD5E-s1229731--99cc64e99df0025ccb0341cd0dcf688b.pdf
│ ├── functional_sub-08.pdf -> ../../.git/annex/objects/MX/vQ/MD5E-s1222308--e13c56f17109d3f142c9c4db60fea674.pdf/MD5E-s1222308--e13c56f17109d3f142c9c4db60fea674.pdf
│ ├── functional_sub-09.pdf -> ../../.git/annex/objects/90/0F/MD5E-s1265097--41a69211a0569413917ce3825eac95d6.pdf/MD5E-s1265097--41a69211a0569413917ce3825eac95d6.pdf
│ ├── functional_sub-10.pdf -> ../../.git/annex/objects/FZ/gq/MD5E-s1299358--12ccfc4a5f52b077b99481fe25aa8ef1.pdf/MD5E-s1299358--12ccfc4a5f52b077b99481fe25aa8ef1.pdf
│ ├── functional_sub-11.pdf -> ../../.git/annex/objects/MJ/mQ/MD5E-s1166014--3465ef6b18514d3cd361c0bffe2b73fc.pdf/MD5E-s1166014--3465ef6b18514d3cd361c0bffe2b73fc.pdf
│ ├── functional_sub-12.pdf -> ../../.git/annex/objects/xp/1f/MD5E-s1177325--6fe4937d5aa567fb5b3c3977362fc9af.pdf/MD5E-s1177325--6fe4937d5aa567fb5b3c3977362fc9af.pdf
│ ├── functional_sub-13.pdf -> ../../.git/annex/objects/4g/vW/MD5E-s1178873--96f341322d21e2bdeb709edc5b047df0.pdf/MD5E-s1178873--96f341322d21e2bdeb709edc5b047df0.pdf
│ ├── functional_sub-14.pdf -> ../../.git/annex/objects/5p/6X/MD5E-s1206987--729f64cf514c9103556c53ccb5430bc4.pdf/MD5E-s1206987--729f64cf514c9103556c53ccb5430bc4.pdf
│ ├── functional_sub-15.pdf -> ../../.git/annex/objects/m1/k9/MD5E-s1223617--9239a1c2d968ed18093b69d28fd9e654.pdf/MD5E-s1223617--9239a1c2d968ed18093b69d28fd9e654.pdf
│ ├── functional_sub-16.pdf -> ../../.git/annex/objects/jq/wP/MD5E-s1294856--5eb7ec97924a22c7e68fd95373694e7e.pdf/MD5E-s1294856--5eb7ec97924a22c7e68fd95373694e7e.pdf
│ ├── functional_sub-17.pdf -> ../../.git/annex/objects/0m/3Q/MD5E-s1238563--129db424a50b7889278024828c08c736.pdf/MD5E-s1238563--129db424a50b7889278024828c08c736.pdf
│ ├── functional_sub-18.pdf -> ../../.git/annex/objects/VF/Fm/MD5E-s1197868--3b23e8d53b11d98d49b1adf62ff559df.pdf/MD5E-s1197868--3b23e8d53b11d98d49b1adf62ff559df.pdf
│ ├── functional_sub-19.pdf -> ../../.git/annex/objects/Jj/m8/MD5E-s1164028--aea7dfa78e9be2e83a9b313f2ebdc4bd.pdf/MD5E-s1164028--aea7dfa78e9be2e83a9b313f2ebdc4bd.pdf
│ ├── functional_sub-20.pdf -> ../../.git/annex/objects/x1/ZQ/MD5E-s1292308--8869b1b640797a2be2aa03be69b89840.pdf/MD5E-s1292308--8869b1b640797a2be2aa03be69b89840.pdf
│ ├── functional_sub-21.pdf -> ../../.git/annex/objects/MG/zW/MD5E-s1216271--3d5c5ca0f8f4ba06b3289e197a40defd.pdf/MD5E-s1216271--3d5c5ca0f8f4ba06b3289e197a40defd.pdf
│ ├── functional_sub-22.pdf -> ../../.git/annex/objects/5m/pj/MD5E-s1142289--4f9e57d8bfe3d39881c43b959189d69f.pdf/MD5E-s1142289--4f9e57d8bfe3d39881c43b959189d69f.pdf
│ ├── functional_sub-23.pdf -> ../../.git/annex/objects/m7/Z2/MD5E-s1233046--7758914aecbf2b5d01cd0825952609be.pdf/MD5E-s1233046--7758914aecbf2b5d01cd0825952609be.pdf
│ ├── functional_sub-24.pdf -> ../../.git/annex/objects/mz/5m/MD5E-s1265224--c188bd88fc1c99308389f528ea4df71e.pdf/MD5E-s1265224--c188bd88fc1c99308389f528ea4df71e.pdf
│ ├── functional_sub-25.pdf -> ../../.git/annex/objects/Mk/G6/MD5E-s1260984--1b16abcbbf55ccc7763f1d704d76628f.pdf/MD5E-s1260984--1b16abcbbf55ccc7763f1d704d76628f.pdf
│ └── functional_sub-26.pdf -> ../../.git/annex/objects/1m/zq/MD5E-s1285726--6838f727d5c4b5593a7b5e0e6b20483a.pdf/MD5E-s1285726--6838f727d5c4b5593a7b5e0e6b20483a.pdf
├── participants.tsv
├── sub-01
│ ├── anat
│ │ └── sub-01_T1w.nii.gz -> ../../.git/annex/objects/Pf/6k/MD5E-s10581116--757e697a01eeea5c97a7d6fbc7153373.nii.gz/MD5E-s10581116--757e697a01eeea5c97a7d6fbc7153373.nii.gz
│ └── func
│ ├── sub-01_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/5m/w9/MD5E-s28061534--8e8c44ff53f9b5d46f2caae5916fa4ef.nii.gz/MD5E-s28061534--8e8c44ff53f9b5d46f2caae5916fa4ef.nii.gz
│ ├── sub-01_task-flanker_run-1_events.tsv
│ ├── sub-01_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/2F/58/MD5E-s28143286--f0bcf782c3688e2cf7149b4665949484.nii.gz/MD5E-s28143286--f0bcf782c3688e2cf7149b4665949484.nii.gz
│ └── sub-01_task-flanker_run-2_events.tsv
├── sub-02
│ ├── anat
│ │ └── sub-02_T1w.nii.gz -> ../../.git/annex/objects/3m/FF/MD5E-s10737123--cbd4181ee26559e8ec0a441fa2f834a7.nii.gz/MD5E-s10737123--cbd4181ee26559e8ec0a441fa2f834a7.nii.gz
│ └── func
│ ├── sub-02_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/8v/2j/MD5E-s29188378--80050f0deb13562c24f2fc23f8d095bd.nii.gz/MD5E-s29188378--80050f0deb13562c24f2fc23f8d095bd.nii.gz
│ ├── sub-02_task-flanker_run-1_events.tsv
│ ├── sub-02_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/fM/Kw/MD5E-s29193540--cc013f2d7d148b448edca8aada349d02.nii.gz/MD5E-s29193540--cc013f2d7d148b448edca8aada349d02.nii.gz
│ └── sub-02_task-flanker_run-2_events.tsv
├── sub-03
│ ├── anat
│ │ └── sub-03_T1w.nii.gz -> ../../.git/annex/objects/7W/9z/MD5E-s10707026--8f1858934cc7c7457e3a4a71cc2131fc.nii.gz/MD5E-s10707026--8f1858934cc7c7457e3a4a71cc2131fc.nii.gz
│ └── func
│ ├── sub-03_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/q6/kF/MD5E-s28755729--b19466702eee6b9385bd6e19e362f94c.nii.gz/MD5E-s28755729--b19466702eee6b9385bd6e19e362f94c.nii.gz
│ ├── sub-03_task-flanker_run-1_events.tsv
│ ├── sub-03_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/zV/K1/MD5E-s28782544--8d9700a435d08c90f0c1d534efdc8b69.nii.gz/MD5E-s28782544--8d9700a435d08c90f0c1d534efdc8b69.nii.gz
│ └── sub-03_task-flanker_run-2_events.tsv
├── sub-04
│ ├── anat
│ │ └── sub-04_T1w.nii.gz -> ../../.git/annex/objects/FW/14/MD5E-s10738444--2a9a2ba4ea7d2324c84bf5a2882f196c.nii.gz/MD5E-s10738444--2a9a2ba4ea7d2324c84bf5a2882f196c.nii.gz
│ └── func
│ ├── sub-04_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/9Z/0Q/MD5E-s29062799--27171406951ea275cb5857ea0dc32345.nii.gz/MD5E-s29062799--27171406951ea275cb5857ea0dc32345.nii.gz
│ ├── sub-04_task-flanker_run-1_events.tsv
│ ├── sub-04_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/FW/FZ/MD5E-s29071279--f89b61fe3ebab26df1374f2564bd95c2.nii.gz/MD5E-s29071279--f89b61fe3ebab26df1374f2564bd95c2.nii.gz
│ └── sub-04_task-flanker_run-2_events.tsv
├── sub-05
│ ├── anat
│ │ └── sub-05_T1w.nii.gz -> ../../.git/annex/objects/k2/Kj/MD5E-s10753867--c4b5788da5f4c627f0f5862da5f46c35.nii.gz/MD5E-s10753867--c4b5788da5f4c627f0f5862da5f46c35.nii.gz
│ └── func
│ ├── sub-05_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/VZ/z5/MD5E-s29667270--0ce9ac78b6aa9a77fc94c655a6ff5a06.nii.gz/MD5E-s29667270--0ce9ac78b6aa9a77fc94c655a6ff5a06.nii.gz
│ ├── sub-05_task-flanker_run-1_events.tsv
│ ├── sub-05_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/z7/MP/MD5E-s29660544--752750dabb21e2cf28e87d1d550a71b9.nii.gz/MD5E-s29660544--752750dabb21e2cf28e87d1d550a71b9.nii.gz
│ └── sub-05_task-flanker_run-2_events.tsv
├── sub-06
│ ├── anat
│ │ └── sub-06_T1w.nii.gz -> ../../.git/annex/objects/5w/G0/MD5E-s10620585--1132eab3830fe59b8a10b6582bb49004.nii.gz/MD5E-s10620585--1132eab3830fe59b8a10b6582bb49004.nii.gz
│ └── func
│ ├── sub-06_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/3x/qj/MD5E-s29386982--e671c0c647ce7d0d4596e35b702ee970.nii.gz/MD5E-s29386982--e671c0c647ce7d0d4596e35b702ee970.nii.gz
│ ├── sub-06_task-flanker_run-1_events.tsv
│ ├── sub-06_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/9j/6P/MD5E-s29379265--e513a2746d2b5c603f96044cf48c557c.nii.gz/MD5E-s29379265--e513a2746d2b5c603f96044cf48c557c.nii.gz
│ └── sub-06_task-flanker_run-2_events.tsv
├── sub-07
│ ├── anat
│ │ └── sub-07_T1w.nii.gz -> ../../.git/annex/objects/08/fF/MD5E-s10718092--38481fbc489dfb1ec4b174b57591a074.nii.gz/MD5E-s10718092--38481fbc489dfb1ec4b174b57591a074.nii.gz
│ └── func
│ ├── sub-07_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/z1/7W/MD5E-s28946009--5baf7a314874b280543fc0f91f2731af.nii.gz/MD5E-s28946009--5baf7a314874b280543fc0f91f2731af.nii.gz
│ ├── sub-07_task-flanker_run-1_events.tsv
│ ├── sub-07_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/Jf/W7/MD5E-s28960603--682e13963bfc49cc6ae05e9ba5c62619.nii.gz/MD5E-s28960603--682e13963bfc49cc6ae05e9ba5c62619.nii.gz
│ └── sub-07_task-flanker_run-2_events.tsv
├── sub-08
│ ├── anat
│ │ └── sub-08_T1w.nii.gz -> ../../.git/annex/objects/mw/MM/MD5E-s10561256--b94dddd8dc1c146aa8cd97f8d9994146.nii.gz/MD5E-s10561256--b94dddd8dc1c146aa8cd97f8d9994146.nii.gz
│ └── func
│ ├── sub-08_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/zX/v9/MD5E-s28641609--47314e6d1a14b8545686110b5b67f8b8.nii.gz/MD5E-s28641609--47314e6d1a14b8545686110b5b67f8b8.nii.gz
│ ├── sub-08_task-flanker_run-1_events.tsv
│ ├── sub-08_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/WZ/F0/MD5E-s28636310--4535bf26281e1c5556ad0d3468e7fe4e.nii.gz/MD5E-s28636310--4535bf26281e1c5556ad0d3468e7fe4e.nii.gz
│ └── sub-08_task-flanker_run-2_events.tsv
├── sub-09
│ ├── anat
│ │ └── sub-09_T1w.nii.gz -> ../../.git/annex/objects/QJ/ZZ/MD5E-s10775967--e6a18e64bc0a6b17254a9564cf9b8f82.nii.gz/MD5E-s10775967--e6a18e64bc0a6b17254a9564cf9b8f82.nii.gz
│ └── func
│ ├── sub-09_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/k9/1X/MD5E-s29200533--59e86a903e0ab3d1d320c794ba1f0777.nii.gz/MD5E-s29200533--59e86a903e0ab3d1d320c794ba1f0777.nii.gz
│ ├── sub-09_task-flanker_run-1_events.tsv
│ ├── sub-09_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/W3/94/MD5E-s29223017--7f3fb9e260d3bd28e29b0b586ce4c344.nii.gz/MD5E-s29223017--7f3fb9e260d3bd28e29b0b586ce4c344.nii.gz
│ └── sub-09_task-flanker_run-2_events.tsv
├── sub-10
│ ├── anat
│ │ └── sub-10_T1w.nii.gz -> ../../.git/annex/objects/5F/3f/MD5E-s10750712--bde2309077bffe22cb65e42ebdce5bfa.nii.gz/MD5E-s10750712--bde2309077bffe22cb65e42ebdce5bfa.nii.gz
│ └── func
│ ├── sub-10_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/3p/qp/MD5E-s29732696--339715d5cec387f4d44dfe94f304a429.nii.gz/MD5E-s29732696--339715d5cec387f4d44dfe94f304a429.nii.gz
│ ├── sub-10_task-flanker_run-1_events.tsv
│ ├── sub-10_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/11/Zx/MD5E-s29724034--16f2bf452524a315182f188becc1866d.nii.gz/MD5E-s29724034--16f2bf452524a315182f188becc1866d.nii.gz
│ └── sub-10_task-flanker_run-2_events.tsv
├── sub-11
│ ├── anat
│ │ └── sub-11_T1w.nii.gz -> ../../.git/annex/objects/kj/xX/MD5E-s10534963--9e5bff7ec0b5df2850e1d05b1af281ba.nii.gz/MD5E-s10534963--9e5bff7ec0b5df2850e1d05b1af281ba.nii.gz
│ └── func
│ ├── sub-11_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/35/fk/MD5E-s28226875--d5012074c2c7a0a394861b010bcf9a8f.nii.gz/MD5E-s28226875--d5012074c2c7a0a394861b010bcf9a8f.nii.gz
│ ├── sub-11_task-flanker_run-1_events.tsv
│ ├── sub-11_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/j7/ff/MD5E-s28198976--c0a64e3b549568c44bb40b1588027c9a.nii.gz/MD5E-s28198976--c0a64e3b549568c44bb40b1588027c9a.nii.gz
│ └── sub-11_task-flanker_run-2_events.tsv
├── sub-12
│ ├── anat
│ │ └── sub-12_T1w.nii.gz -> ../../.git/annex/objects/kx/2F/MD5E-s10550168--a7f651adc817b6678148b575654532a4.nii.gz/MD5E-s10550168--a7f651adc817b6678148b575654532a4.nii.gz
│ └── func
│ ├── sub-12_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/M0/fX/MD5E-s28403807--f1c3eb2e519020f4315a696ea845fc01.nii.gz/MD5E-s28403807--f1c3eb2e519020f4315a696ea845fc01.nii.gz
│ ├── sub-12_task-flanker_run-1_events.tsv
│ ├── sub-12_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/vW/V0/MD5E-s28424992--8740628349be3c056a0411bf4a852b25.nii.gz/MD5E-s28424992--8740628349be3c056a0411bf4a852b25.nii.gz
│ └── sub-12_task-flanker_run-2_events.tsv
├── sub-13
│ ├── anat
│ │ └── sub-13_T1w.nii.gz -> ../../.git/annex/objects/wM/Xw/MD5E-s10609761--440413c3251d182086105649164222c6.nii.gz/MD5E-s10609761--440413c3251d182086105649164222c6.nii.gz
│ └── func
│ ├── sub-13_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/mf/M4/MD5E-s28180916--aa35f4ad0cf630d6396a8a2dd1f3dda6.nii.gz/MD5E-s28180916--aa35f4ad0cf630d6396a8a2dd1f3dda6.nii.gz
│ ├── sub-13_task-flanker_run-1_events.tsv
│ ├── sub-13_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/XP/76/MD5E-s28202786--8caf1ac548c87b2b35f85e8ae2bf72c1.nii.gz/MD5E-s28202786--8caf1ac548c87b2b35f85e8ae2bf72c1.nii.gz
│ └── sub-13_task-flanker_run-2_events.tsv
├── sub-14
│ ├── anat
│ │ └── sub-14_T1w.nii.gz -> ../../.git/annex/objects/Zw/0z/MD5E-s9223596--33abfb5da565f3487e3a7aebc15f940c.nii.gz/MD5E-s9223596--33abfb5da565f3487e3a7aebc15f940c.nii.gz
│ └── func
│ ├── sub-14_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/Jp/29/MD5E-s29001492--250f1e4daa9be1d95e06af0d56629cc9.nii.gz/MD5E-s29001492--250f1e4daa9be1d95e06af0d56629cc9.nii.gz
│ ├── sub-14_task-flanker_run-1_events.tsv
│ ├── sub-14_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/PK/V2/MD5E-s29068193--5621a3b0af8132c509420b4ad9aaf8fb.nii.gz/MD5E-s29068193--5621a3b0af8132c509420b4ad9aaf8fb.nii.gz
│ └── sub-14_task-flanker_run-2_events.tsv
├── sub-15
│ ├── anat
│ │ └── sub-15_T1w.nii.gz -> ../../.git/annex/objects/Mz/qq/MD5E-s10752891--ddd2622f115ec0d29a0c7ab2366f6f95.nii.gz/MD5E-s10752891--ddd2622f115ec0d29a0c7ab2366f6f95.nii.gz
│ └── func
│ ├── sub-15_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/08/JJ/MD5E-s28285239--feda22c4526af1910fcee58d4c42f07e.nii.gz/MD5E-s28285239--feda22c4526af1910fcee58d4c42f07e.nii.gz
│ ├── sub-15_task-flanker_run-1_events.tsv
│ ├── sub-15_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/9f/0W/MD5E-s28289760--433000a1def662e72d8433dba151c61b.nii.gz/MD5E-s28289760--433000a1def662e72d8433dba151c61b.nii.gz
│ └── sub-15_task-flanker_run-2_events.tsv
├── sub-16
│ ├── anat
│ │ └── sub-16_T1w.nii.gz -> ../../.git/annex/objects/4g/8k/MD5E-s10927450--a196f7075c793328dd6ff3cebf36ea6b.nii.gz/MD5E-s10927450--a196f7075c793328dd6ff3cebf36ea6b.nii.gz
│ └── func
│ ├── sub-16_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/9z/g2/MD5E-s29757991--1a1648b2fa6cc74e31c94f109d8137ba.nii.gz/MD5E-s29757991--1a1648b2fa6cc74e31c94f109d8137ba.nii.gz
│ ├── sub-16_task-flanker_run-1_events.tsv
│ ├── sub-16_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/k8/4F/MD5E-s29773832--fe08739ea816254395b985ee704aaa99.nii.gz/MD5E-s29773832--fe08739ea816254395b985ee704aaa99.nii.gz
│ └── sub-16_task-flanker_run-2_events.tsv
├── sub-17
│ ├── anat
│ │ └── sub-17_T1w.nii.gz -> ../../.git/annex/objects/jQ/MQ/MD5E-s10826014--8e2a6b062df4d1c4327802f2b905ef36.nii.gz/MD5E-s10826014--8e2a6b062df4d1c4327802f2b905ef36.nii.gz
│ └── func
│ ├── sub-17_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/Wz/2P/MD5E-s28991563--9845f461a017a39d1f6e18baaa0c9c41.nii.gz/MD5E-s28991563--9845f461a017a39d1f6e18baaa0c9c41.nii.gz
│ ├── sub-17_task-flanker_run-1_events.tsv
│ ├── sub-17_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/jF/3m/MD5E-s29057821--84ccc041163bcc5b3a9443951e2a5a78.nii.gz/MD5E-s29057821--84ccc041163bcc5b3a9443951e2a5a78.nii.gz
│ └── sub-17_task-flanker_run-2_events.tsv
├── sub-18
│ ├── anat
│ │ └── sub-18_T1w.nii.gz -> ../../.git/annex/objects/3v/pK/MD5E-s10571510--6fc4b5792bc50ea4d14eb5247676fafe.nii.gz/MD5E-s10571510--6fc4b5792bc50ea4d14eb5247676fafe.nii.gz
│ └── func
│ ├── sub-18_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/94/P2/MD5E-s28185776--5b3879ec6fc4bbe1e48efc64984f88cf.nii.gz/MD5E-s28185776--5b3879ec6fc4bbe1e48efc64984f88cf.nii.gz
│ ├── sub-18_task-flanker_run-1_events.tsv
│ ├── sub-18_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/qp/6K/MD5E-s28234699--58019d798a133e5d7806569374dd8160.nii.gz/MD5E-s28234699--58019d798a133e5d7806569374dd8160.nii.gz
│ └── sub-18_task-flanker_run-2_events.tsv
├── sub-19
│ ├── anat
│ │ └── sub-19_T1w.nii.gz -> ../../.git/annex/objects/Zw/p8/MD5E-s8861893--d338005753d8af3f3d7bd8dc293e2a97.nii.gz/MD5E-s8861893--d338005753d8af3f3d7bd8dc293e2a97.nii.gz
│ └── func
│ ├── sub-19_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/04/k6/MD5E-s28178448--3874e748258cf19aa69a05a7c37ad137.nii.gz/MD5E-s28178448--3874e748258cf19aa69a05a7c37ad137.nii.gz
│ ├── sub-19_task-flanker_run-1_events.tsv
│ ├── sub-19_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/mz/P4/MD5E-s28190932--91e6b3e4318ca28f01de8cb967cf8421.nii.gz/MD5E-s28190932--91e6b3e4318ca28f01de8cb967cf8421.nii.gz
│ └── sub-19_task-flanker_run-2_events.tsv
├── sub-20
│ ├── anat
│ │ └── sub-20_T1w.nii.gz -> ../../.git/annex/objects/g1/FF/MD5E-s11025608--5929806a7aa5720fc755687e1450b06c.nii.gz/MD5E-s11025608--5929806a7aa5720fc755687e1450b06c.nii.gz
│ └── func
│ ├── sub-20_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/v5/ZJ/MD5E-s29931631--bf9abb057367ce66961f0b7913e8e707.nii.gz/MD5E-s29931631--bf9abb057367ce66961f0b7913e8e707.nii.gz
│ ├── sub-20_task-flanker_run-1_events.tsv
│ ├── sub-20_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/J3/KW/MD5E-s29945590--96cfd5b77cd096f6c6a3530015fea32d.nii.gz/MD5E-s29945590--96cfd5b77cd096f6c6a3530015fea32d.nii.gz
│ └── sub-20_task-flanker_run-2_events.tsv
├── sub-21
│ ├── anat
│ │ └── sub-21_T1w.nii.gz -> ../../.git/annex/objects/K6/6K/MD5E-s8662805--77b262ddd929fa08d78591bfbe558ac6.nii.gz/MD5E-s8662805--77b262ddd929fa08d78591bfbe558ac6.nii.gz
│ └── func
│ ├── sub-21_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/Wz/p9/MD5E-s28756041--9ae556d4e3042532d25af5dc4ab31840.nii.gz/MD5E-s28756041--9ae556d4e3042532d25af5dc4ab31840.nii.gz
│ ├── sub-21_task-flanker_run-1_events.tsv
│ ├── sub-21_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/xF/M3/MD5E-s28758438--81866411fc6b6333ec382a20ff0be718.nii.gz/MD5E-s28758438--81866411fc6b6333ec382a20ff0be718.nii.gz
│ └── sub-21_task-flanker_run-2_events.tsv
├── sub-22
│ ├── anat
│ │ └── sub-22_T1w.nii.gz -> ../../.git/annex/objects/JG/ZV/MD5E-s9282392--9e7296a6a5b68df46b77836182b6681a.nii.gz/MD5E-s9282392--9e7296a6a5b68df46b77836182b6681a.nii.gz
│ └── func
│ ├── sub-22_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/qW/Gw/MD5E-s28002098--c6bea10177a38667ceea3261a642b3c6.nii.gz/MD5E-s28002098--c6bea10177a38667ceea3261a642b3c6.nii.gz
│ ├── sub-22_task-flanker_run-1_events.tsv
│ ├── sub-22_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/VX/Zj/MD5E-s28027568--b34d0df9ad62485aba25296939429885.nii.gz/MD5E-s28027568--b34d0df9ad62485aba25296939429885.nii.gz
│ └── sub-22_task-flanker_run-2_events.tsv
├── sub-23
│ ├── anat
│ │ └── sub-23_T1w.nii.gz -> ../../.git/annex/objects/4Z/4x/MD5E-s10626062--db5a6ba6730b319c6425f2e847ce9b14.nii.gz/MD5E-s10626062--db5a6ba6730b319c6425f2e847ce9b14.nii.gz
│ └── func
│ ├── sub-23_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/VK/8F/MD5E-s28965005--4a9a96d9322563510ca14439e7fd6cea.nii.gz/MD5E-s28965005--4a9a96d9322563510ca14439e7fd6cea.nii.gz
│ ├── sub-23_task-flanker_run-1_events.tsv
│ ├── sub-23_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/56/20/MD5E-s29050413--753b0d2c23c4af6592501219c2e2c6bd.nii.gz/MD5E-s29050413--753b0d2c23c4af6592501219c2e2c6bd.nii.gz
│ └── sub-23_task-flanker_run-2_events.tsv
├── sub-24
│ ├── anat
│ │ └── sub-24_T1w.nii.gz -> ../../.git/annex/objects/jQ/fV/MD5E-s10739691--458f0046eff18ee8c43456637766a819.nii.gz/MD5E-s10739691--458f0046eff18ee8c43456637766a819.nii.gz
│ └── func
│ ├── sub-24_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/km/fV/MD5E-s29354610--29ebfa60e52d49f7dac6814cb5fdc2bc.nii.gz/MD5E-s29354610--29ebfa60e52d49f7dac6814cb5fdc2bc.nii.gz
│ ├── sub-24_task-flanker_run-1_events.tsv
│ ├── sub-24_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/Wj/KK/MD5E-s29423307--fedaa1d7c6e34420735bb3bbe5a2fe38.nii.gz/MD5E-s29423307--fedaa1d7c6e34420735bb3bbe5a2fe38.nii.gz
│ └── sub-24_task-flanker_run-2_events.tsv
├── sub-25
│ ├── anat
│ │ └── sub-25_T1w.nii.gz -> ../../.git/annex/objects/Gk/FQ/MD5E-s8998578--f560d832f13e757b485c16d570bf6ebc.nii.gz/MD5E-s8998578--f560d832f13e757b485c16d570bf6ebc.nii.gz
│ └── func
│ ├── sub-25_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/XW/1v/MD5E-s29473003--49b04e7e4b450ec5ef93ff02d4158775.nii.gz/MD5E-s29473003--49b04e7e4b450ec5ef93ff02d4158775.nii.gz
│ ├── sub-25_task-flanker_run-1_events.tsv
│ ├── sub-25_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/Qm/M7/MD5E-s29460132--b0e9039e9f33510631f229c8c2193285.nii.gz/MD5E-s29460132--b0e9039e9f33510631f229c8c2193285.nii.gz
│ └── sub-25_task-flanker_run-2_events.tsv
├── sub-26
│ ├── anat
│ │ └── sub-26_T1w.nii.gz -> ../../.git/annex/objects/kf/9F/MD5E-s10850250--5f103b2660f488e4afa193f9307c1291.nii.gz/MD5E-s10850250--5f103b2660f488e4afa193f9307c1291.nii.gz
│ └── func
│ ├── sub-26_task-flanker_run-1_bold.nii.gz -> ../../.git/annex/objects/QV/10/MD5E-s30127491--8e30aa4bbfcc461bac8598bf621283c5.nii.gz/MD5E-s30127491--8e30aa4bbfcc461bac8598bf621283c5.nii.gz
│ ├── sub-26_task-flanker_run-1_events.tsv
│ ├── sub-26_task-flanker_run-2_bold.nii.gz -> ../../.git/annex/objects/3G/Q6/MD5E-s30162480--80fd132e7cb1600ab248249e78f6f1aa.nii.gz/MD5E-s30162480--80fd132e7cb1600ab248249e78f6f1aa.nii.gz
│ └── sub-26_task-flanker_run-2_events.tsv
└── task-flanker_bold.json
80 directories, 192 files
Inspecting the anatomical and functional images#
To use the AFNI graphical user interface, you would type:
afni sub-08_T1w.nii.gz
We will use ipyniivue to look at the data:
Many of the quality checks for the functional images are similar to those for the anatomical images. Look out for excessively bright or dark spots in the grey or white matter, as well as any image distortions, such as abnormal stretching or warping. A common area where slight distortion may occur is in the orbitofrontal region, just above the eyeballs.
Additionally, it’s important to check for excessive motion during the scan.
anat_image = './ds000102/sub-08/anat/sub-08_T1w.nii.gz'
func1_image = './ds000102/sub-08/func/sub-08_task-flanker_run-1_bold.nii.gz'
func2_image = './ds000102/sub-08/func/sub-08_task-flanker_run-2_bold.nii.gz'
# anatomical image
nv = NiiVue()
nv.load_volumes([{"path": anat_image}])
nv
Image(url='https://raw.githubusercontent.com/NeuroDesk/example-notebooks/refs/heads/main/books/images/afni_preproc_anat.png')
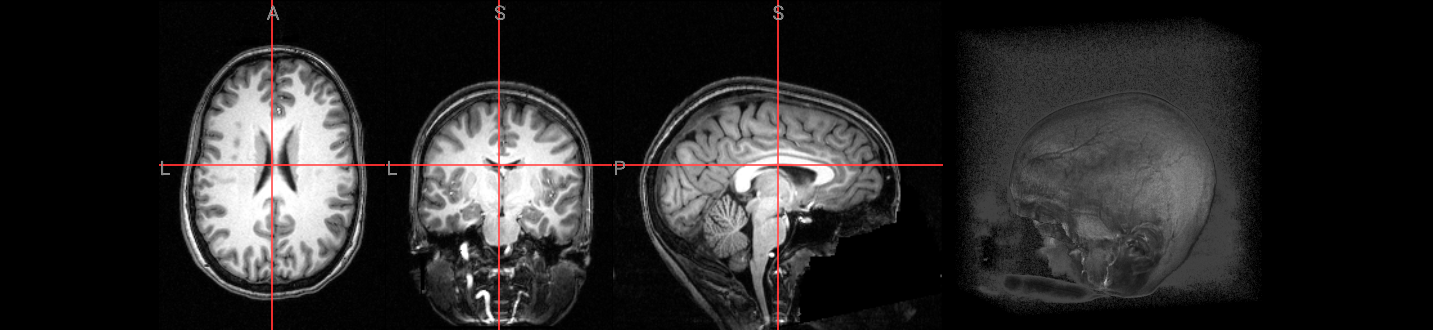
# functional run 1
nv = NiiVue()
nv.load_volumes([{"path": func1_image}])
nv
Image(url='https://raw.githubusercontent.com/NeuroDesk/example-notebooks/refs/heads/main/books/images/afni_preproc_func1.png')
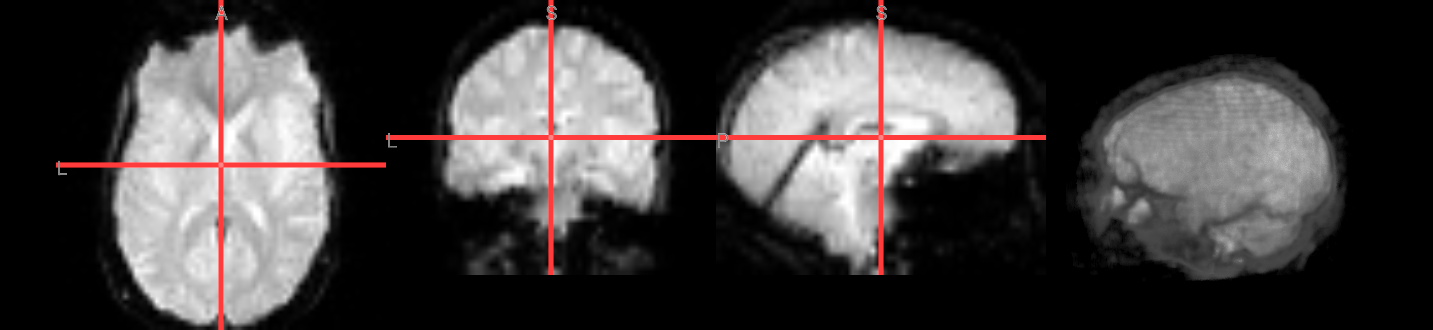
AFNI Commands and Preprocessing#
We will be working with the data from subject sub-08. First, we’ll introduce basic AFNI commands. Then, we’ll create a processing script using afni_proc.py, which automates the entire preprocessing workflow. AFNI also provides graphical user interfaces (GUIs), such as uber_subject.py and uber_ttest.py (for group analysis), which help build processing scripts by combining many commands in the correct order. However, since GUIs cannot be used within a Jupyter Notebook environment, we will focus on writing the scripts directly.
After setting up the automated script, we’ll walk through each preprocessing step individually and discuss how to check data quality before and after each step.
1: AFNI Commands
2: Preprocessing with
afni_prc.pyRegistration and Normalization
Alignment and Motion Correction
Smoothing
Masking and Scaling
Checking Preprocessing
1. AFNI commands#
AFNI commands typicallly require at least one argument, or input, and they also usually require you to specify what to call the output of the command.
Let’s take skull-stripping, for example - a common preprocessing step that removes the skull from the brain. The AFNI command to do this step is called 3dSkullStrip. Use the -h flag to print the help page.
!3dSkullStrip -h
Usage: A program to extract the brain from surrounding.
tissue from MRI T1-weighted images.
The simplest command would be:
3dSkullStrip <-input DSET>
Also consider the script @SSwarper, which combines the use of
3dSkullStrip and nonlinear warping to an MNI template to produce
a skull-stripped dataset in MNI space, plus the nonlinear warp
that can used to transform other datasets from the same subject
(e.g., EPI) to MNI space. (This script only applies to human brain
images.)
The fully automated process consists of three steps:
1- Preprocessing of volume to remove gross spatial image
non-uniformity artifacts and reposition the brain in
a reasonable manner for convenience.
** Note that in many cases, using 3dUnifize before **
** using 3dSkullStrip will give better results. **
2- Expand a spherical surface iteratively until it envelopes
the brain. This is a modified version of the BET algorithm:
Fast robust automated brain extraction,
by Stephen M. Smith, HBM 2002 v 17:3 pp 143-155
Modifications include the use of:
. outer brain surface
. expansion driven by data inside and outside the surface
. avoidance of eyes and ventricles
. a set of operations to avoid the clipping of certain brain
areas and reduce leakage into the skull in heavily shaded
data
. two additional processing stages to ensure convergence and
reduction of clipped areas.
. use of 3d edge detection, see Deriche and Monga references
in 3dedge3 -help.
3- The creation of various masks and surfaces modeling brain
and portions of the skull
Common examples of usage:
-------------------------
o 3dSkullStrip -input VOL -prefix VOL_PREFIX
Vanilla mode, should work for most datasets.
o 3dSkullStrip -input VOL -prefix VOL_PREFIX -push_to_edge
Adds an agressive push to brain edges. Use this option
when the chunks of gray matter are not included. This option
might cause the mask to leak into non-brain areas.
o 3dSkullStrip -input VOL -surface_coil -prefix VOL_PREFIX -monkey
Vanilla mode, for use with monkey data.
o 3dSkullStrip -input VOL -prefix VOL_PREFIX -ld 30
Use a denser mesh, in the cases where you have lots of
csf between gyri. Also helps when some of the brain is clipped
close to regions of high curvature.
Tips:
-----
I ran the program with the default parameters on 200+ datasets.
The results were quite good in all but a couple of instances, here
are some tips on fixing trouble spots:
Clipping in frontal areas, close to the eye balls:
+ Try -push_to_edge option first.
Can also try -no_avoid_eyes option.
Clipping in general:
+ Try -push_to_edge option first.
Can also use lower -shrink_fac, start with 0.5 then 0.4
Problems down below:
+ Piece of cerebellum missing, reduce -shrink_fac_bot_lim
from default value.
+ Leakage in lower areas, increase -shrink_fac_bot_lim
from default value.
Some lobules are not included:
+ Use a denser mesh. Start with -ld 30. If that still fails,
try even higher density (like -ld 50) and increase iterations
(say to -niter 750).
Expect the program to take much longer in that case.
+ Instead of using denser meshes, you could try blurring the data
before skull stripping. Something like -blur_fwhm 2 did
wonders for some of my data with the default options of 3dSkullStrip
Blurring is a lot faster than increasing mesh density.
+ Use also a smaller -shrink_fac is you have lots of CSF between
gyri.
Massive chunks missing:
+ If brain has very large ventricles and lots of CSF between gyri,
the ventricles will keep attracting the surface inwards.
This often happens with older brains. In such
cases, use the -visual option to see what is happening.
For example, the options below did the trick in various
instances.
-blur_fwhm 2 -use_skull
or for more stubborn cases increase csf avoidance with this cocktail
-blur_fwhm 2 -use_skull -avoid_vent -avoid_vent -init_radius 75
+ Too much neck in the volume might throw off the initialization
step. You can fix this by clipping tissue below the brain with
@clip_volume -below ZZZ -input INPUT
where ZZZ is a Z coordinate somewhere below the brain.
Large regions outside brain included:
+ Usually because noise level is high. Try @NoisySkullStrip.
Make sure that brain orientation is correct. This means the image in
AFNI's axial slice viewer should be close to the brain's axial plane.
The same goes for the other planes. Otherwise, the program might do a lousy
job removing the skull.
Eye Candy Mode:
---------------
You can run 3dSkullStrip and have it send successive iterations
to SUMA and AFNI. This is very helpful in following the
progression of the algorithm and determining the source
of trouble, if any.
Example:
afni -niml -yesplugouts &
suma -niml &
3dSkullStrip -input Anat+orig -o_ply anat_brain -visual
Help section for the intrepid:
------------------------------
3dSkullStrip < -input VOL >
[< -o_TYPE PREFIX >] [< -prefix VOL_PREFIX >]
[< -spatnorm >] [< -no_spatnorm >] [< -write_spatnorm >]
[< -niter N_ITER >] [< -ld LD >]
[< -shrink_fac SF >] [< -var_shrink_fac >]
[< -no_var_shrink_fac >] [< -shrink_fac_bot_lim SFBL >]
[< -pushout >] [< -no_pushout >] [< -exp_frac FRAC]
[< -touchup >] [< -no_touchup >]
[< -fill_hole R >] [< -NN_smooth NN_SM >]
[< -smooth_final SM >] [< -avoid_vent >] [< -no_avoid_vent >]
[< -use_skull >] [< -no_use_skull >]
[< -avoid_eyes >] [< -no_avoid_eyes >]
[< -use_edge >] [< -no_use_edge >]
[< -push_to_edge >] [<-no_push_to_edge>]
[< -perc_int PERC_INT >]
[< -max_inter_iter MII >] [-mask_vol | -orig_vol | -norm_vol]
[< -debug DBG >] [< -node_debug NODE_DBG >]
[< -demo_pause >]
[< -monkey >] [< -marmoset >] [<-rat>]
NOTE: Please report bugs and strange failures
to saadz@mail.nih.gov
Mandatory parameters:
-input VOL: Input AFNI (or AFNI readable) volume.
Optional Parameters:
-monkey: the brain of a monkey.
-marmoset: the brain of a marmoset.
this one was tested on one dataset
and may not work with non default
options. Check your results!
-rat: the brain of a rat.
By default, no_touchup is used with the rat.
-surface_coil: Data acquired with a surface coil.
-o_TYPE PREFIX: prefix of output surface.
where TYPE specifies the format of the surface
and PREFIX is, well, the prefix.
TYPE is one of: fs, 1d (or vec), sf, ply.
More on that below.
-skulls: Output surface models of the skull.
-4Tom: The output surfaces are named based
on PREFIX following -o_TYPE option below.
-prefix VOL_PREFIX: prefix of output volume.
If not specified, the prefix is the same
as the one used with -o_TYPE.
The output volume is skull stripped version
of the input volume. In the earlier version
of the program, a mask volume was written out.
You can still get that mask volume instead of the
skull-stripped volume with the option -mask_vol .
NOTE: In the default setting, the output volume does not
have values identical to those in the input.
In particular, the range might be larger
and some low-intensity values are set to 0.
If you insist on having the same range of values as in
the input, then either use option -orig_vol, or run:
3dcalc -nscale -a VOL+VIEW -b VOL_PREFIX+VIEW \
-expr 'a*step(b)' -prefix VOL_SAME_RANGE
With the command above, you can preserve the range
of values of the input but some low-intensity voxels would
still be masked. If you want to preserve them, then use
-mask_vol in the 3dSkullStrip command that would produce
VOL_MASK_PREFIX+VIEW. Then run 3dcalc masking with voxels
inside the brain surface envelope:
3dcalc -nscale -a VOL+VIEW -b VOL_MASK_PREFIX+VIEW \
-expr 'a*step(b-3.01)' -prefix VOL_SAME_RANGE_KEEP_LOW
-norm_vol: Output a masked and somewhat intensity normalized and
thresholded version of the input. This is the default,
and you can use -orig_vol to override it.
-orig_vol: Output a masked version of the input AND do not modify
the values inside the brain as -norm_vol would.
-mask_vol: Output a mask volume instead of a skull-stripped
volume.
The mask volume containes:
0: Voxel outside surface
1: Voxel just outside the surface. This means the voxel
center is outside the surface but inside the
bounding box of a triangle in the mesh.
2: Voxel intersects the surface (a triangle), but center
lies outside.
3: Voxel contains a surface node.
4: Voxel intersects the surface (a triangle), center lies
inside surface.
5: Voxel just inside the surface. This means the voxel
center is inside the surface and inside the
bounding box of a triangle in the mesh.
6: Voxel inside the surface.
-spat_norm: (Default) Perform spatial normalization first.
This is a necessary step unless the volume has
been 'spatnormed' already.
-no_spatnorm: Do not perform spatial normalization.
Use this option only when the volume
has been run through the 'spatnorm' process
-spatnorm_dxyz DXYZ: Use DXY for the spatial resolution of the
spatially normalized volume. The default
is the lowest of all three dimensions.
For human brains, use DXYZ of 1.0, for
primate brain, use the default setting.
-write_spatnorm: Write the 'spatnormed' volume to disk.
-niter N_ITER: Number of iterations. Default is 250
For denser meshes, you need more iterations
N_ITER of 750 works for LD of 50.
-ld LD: Parameter to control the density of the surface.
Default is 20 if -no_use_edge is used,
30 with -use_edge. See CreateIcosahedron -help
for details on this option.
-shrink_fac SF: Parameter controlling the brain vs non-brain
intensity threshold (tb). Default is 0.6.
tb = (Imax - t2) SF + t2
where t2 is the 2 percentile value and Imax is the local
maximum, limited to the median intensity value.
For more information on tb, t2, etc. read the BET paper
mentioned above. Note that in 3dSkullStrip, SF can vary across
iterations and might be automatically clipped in certain areas.
SF can vary between 0 and 1.
0: Intensities < median inensity are considered non-brain
1: Intensities < t2 are considered non-brain
-var_shrink_fac: Vary the shrink factor with the number of
iterations. This reduces the likelihood of a surface
getting stuck on large pools of CSF before reaching
the outer surface of the brain. (Default)
-no_var_shrink_fac: Do not use var_shrink_fac.
-shrink_fac_bot_lim SFBL: Do not allow the varying SF to go
below SFBL . Default 0.65, 0.4 when edge detection is used.
This option helps reduce potential for leakage below
the cerebellum.
In certain cases where you have severe non-uniformity resulting
in low signal towards the bottom of the brain, you will need to
reduce this parameter.
-pushout: Consider values above each node in addition to values
below the node when deciding on expansion. (Default)
-no_pushout: Do not use -pushout.
-exp_frac FRAC: Speed of expansion (see BET paper). Default is 0.1.
-touchup: Perform touchup operations at end to include
areas not covered by surface expansion.
Use -touchup -touchup for aggressive makeup.
(Default is -touchup)
-no_touchup: Do not use -touchup
-fill_hole R: Fill small holes that can result from small surface
intersections caused by the touchup operation.
R is the maximum number of pixels on the side of a hole
that can be filled. Big holes are not filled.
If you use -touchup, the default R is 10. Otherwise
the default is 0.
This is a less than elegant solution to the small
intersections which are usually eliminated
automatically.
-NN_smooth NN_SM: Perform Nearest Neighbor coordinate interpolation
every few iterations. Default is 72
-smooth_final SM: Perform final surface smoothing after all iterations.
Default is 20 smoothing iterations.
Smoothing is done using Taubin's method,
see SurfSmooth -help for detail.
-avoid_vent: avoid ventricles. Default.
Use this option twice to make the avoidance more
agressive. That is at times needed with old brains.
-no_avoid_vent: Do not use -avoid_vent.
-init_radius RAD: Use RAD for the initial sphere radius.
For the automatic setting, there is an
upper limit of 100mm for humans.
For older brains with lots of CSF, you
might benefit from forcing the radius
to something like 75mm
-avoid_eyes: avoid eyes. Default
-no_avoid_eyes: Do not use -avoid_eyes.
-use_edge: Use edge detection to reduce leakage into meninges and eyes.
Default.
-no_use_edge: Do no use edges.
-push_to_edge: Perform aggressive push to edge at the end.
This option might cause leakage.
-no_push_to_edge: (Default).
-use_skull: Use outer skull to limit expansion of surface into
the skull due to very strong shading artifacts.
This option is buggy at the moment, use it only
if you have leakage into skull.
-no_use_skull: Do not use -use_skull (Default).
-send_no_skull: Do not send the skull surface to SUMA if you are
using -talk_suma
-perc_int PERC_INT: Percentage of segments allowed to intersect
surface. Ideally this should be 0 (Default).
However, few surfaces might have small stubborn
intersections that produce a few holes.
PERC_INT should be a small number, typically
between 0 and 0.1. A -1 means do not do
any testing for intersection.
-max_inter_iter N_II: Number of iteration to remove intersection
problems. With each iteration, the program
automatically increases the amount of smoothing
to get rid of intersections. Default is 4
-blur_fwhm FWHM: Blur dset after spatial normalization.
Recommended when you have lots of CSF in brain
and when you have protruding gyri (finger like)
Recommended value is 2..4.
-interactive: Make the program stop at various stages in the
segmentation process for a prompt from the user
to continue or skip that stage of processing.
This option is best used in conjunction with options
-talk_suma and -feed_afni
-demo_pause: Pause at various step in the process to facilitate
interactive demo while 3dSkullStrip is communicating
with AFNI and SUMA. See 'Eye Candy' mode below and
-talk_suma option.
-fac FAC: Multiply input dataset by FAC if range of values is too
small.
-visual: Equivalent to using -talk_suma -feed_afni -send_kth 5
-debug DBG: debug levels of 0 (default), 1, 2, 3.
This is no Rick Reynolds debug, which is oft nicer
than the results, but it will do.
-node_debug NODE_DBG: Output lots of parameters for node
NODE_DBG for each iteration.
The next 3 options are for specifying surface coordinates
to keep the program from having to recompute them.
The options are only useful for saving time during debugging.
-brain_contour_xyz_file BRAIN_CONTOUR_XYZ.1D
-brain_hull_xyz_file BRAIN_HULL_XYZ.1D
-skull_outer_xyz_file SKULL_OUTER_XYZ.1D
-help: The help you need
Compile Date:
Jul 8 2021
Ziad S. Saad SSCC/NIMH/NIH saadz@mail.nih.gov
One of AFNI’s major advantages is the quality of its documentation and help resources. Each command’s usage is thoroughly described, and the purpose behind various options is clearly explained. Example commands are provided to illustrate how to handle different situations — for instance, if too much skull remains after skull-stripping, you might be advised to use a flag like -push_to_edge.
The simplest way to run 3dSkullStrip is by using the -input option to specify the anatomical dataset for processing. The -prefix option is also used to output a NIfTI image for visualization with ipyniivue.
!3dSkullStrip -input $anat_image -prefix anat_ss.nii.gz
The intensity in the output dataset is a modified version
of the intensity in the input volume.
To obtain a masked version of the input with identical values inside
the brain, you can either use 3dSkullStrip's -orig_vol option
or run the following command:
3dcalc -a ./ds000102/sub-08/anat/sub-08_T1w.nii.gz -b ./anat_ss.nii.gz+orig -expr 'a*step(b)' \
-prefix ./anat_ss.nii.gz_orig_vol
to generate a new masked version of the input.
volumes = [{"path": anat_image},
{"path": "./anat_ss.nii.gz", "colormap": "red"}]
nv = NiiVue()
nv.load_volumes(volumes)
nv
Image(url='https://raw.githubusercontent.com/NeuroDesk/example-notebooks/refs/heads/main/books/images/afni_preproc_anat_ss.png')
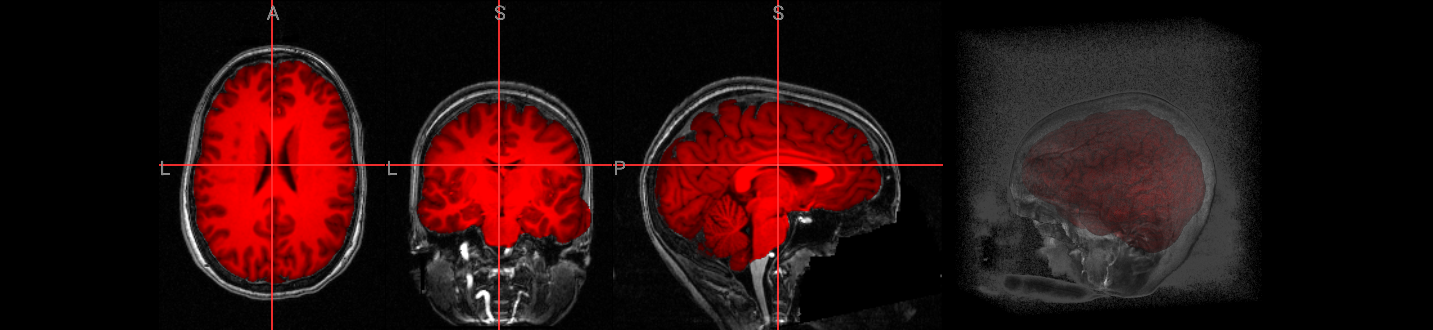
2. Preprocessing#
To automate preprocessing in AFNI, the powerful afni_proc.py tool is used. This command generates a fully customizable tcsh script that includes all necessary preprocessing steps, from slice timing correction to scaling.
The basic idea is:
Specify which processing blocks to apply.
afni_proc.pywrites a script to carry them out in the correct order.The generated script can be reviewed and run, or modified if needed.
The processing blocks include both automatic steps (such as setup and initial time concatenation) and default blocks that you can customize, skip, or reorder. For example:
Automatic blocks:
-setup: Set up subject information, create output directory (subj_id, script, out_dir)
-tcat: Remove unwanted initial TRs.
Default blocks:
-tshift: Slice timing correction.
-volreg: Volume registration (motion correction).
-blur: Spatial smoothing.
-mask: Create a brain mask from EPI data.
-scale: Normalize voxel-wise signal intensities.
-regress: (optional) Regression analysis for task-based designs.
A full list of options and help can be found in the AFNI documentation.
The following command specifies key preprocessing steps for sub-08, including motion correction, normalization to MNI space, smoothing, and scaling. Additionally, the alignment option -giant_move is included, which allows for larger shifts and rotations during alignments.
➡️
setup ➡️ tcat ➡️ tshift ➡️ align ➡️ tlrc ➡️ volreg ➡️ blur ➡️ mask ➡️ scale ➡️ ✅ outputs
!afni_proc.py \
-subj_id sub_08 \
-script proc.sub_08 \
-scr_overwrite \
-out_dir ./afni_processing/sub_08.results \
-blocks tshift align tlrc volreg blur mask scale \
-copy_anat $anat_image \
-dsets \
$func1_image \
$func2_image \
-tcat_remove_first_trs 0 \
-align_opts_aea -giant_move \
-tlrc_base MNI_avg152T1+tlrc \
-volreg_align_to MIN_OUTLIER \
-volreg_align_e2a \
-volreg_tlrc_warp \
-blur_size 4.0
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
-- applying input view as +orig
-- will use min outlier volume as motion base
-- tcat: reps is now 146
++ updating polort to 2, from run len 292.0 s
-- volreg: using base dset vr_base_min_outlier+orig
++ volreg: applying volreg/epi2anat/tlrc xforms to isotropic 3 mm tlrc voxels
-- applying anat warps to 1 dataset(s): sub-08_T1w
-- masking: group anat = 'MNI_avg152T1+tlrc', exists = 1
-- have 1 ROI dict entries ...
-- no regress block, skipping gen_ss_review_scripts.py
-- using default: will not apply EPI Automask
(see 'MASKING NOTE' from the -help for details)
--> script is file: proc.sub_08
to execute via tcsh:
tcsh -xef proc.sub_08 |& tee output.proc.sub_08
to execute via bash:
tcsh -xef proc.sub_08 2>&1 | tee output.proc.sub_08
Run the Preprocessing Script and Inspect the Output
Now that we have generated our preprocessing script proc.sub_08, we can execute it via tcsh and save a full record of the output in a file called output.proc.sub_08.
! tcsh -xef proc.sub_08 |& tee output.proc.sub_08
echo auto-generated by afni_proc.py, Tue Nov 4 03:31:50 2025
auto-generated by afni_proc.py, Tue Nov 4 03:31:50 2025
echo (version 7.16, May 19, 2021)
(version 7.16, May 19, 2021)
echo execution started: `date`
date
execution started: Tue Nov 4 03:31:52 UTC 2025
afni -ver
Precompiled binary linux_openmp_64: Jul 8 2021 (Version AFNI_21.2.00 'Nerva')
afni_history -check_date 27 Jun 2019
-- is current: afni_history as new as: 27 Jun 2019
most recent entry is: 30 Jun 2021
if ( 0 ) then
if ( 0 > 0 ) then
set subj = sub_08
endif
set output_dir = ./afni_processing/sub_08.results
if ( -d ./afni_processing/sub_08.results ) then
set runs = ( `count -digits 2 1 2` )
count -digits 2 1 2
mkdir -p ./afni_processing/sub_08.results
mkdir ./afni_processing/sub_08.results/stimuli
3dcopy ds000102/sub-08/anat/sub-08_T1w.nii.gz ./afni_processing/sub_08.results/sub-08_T1w
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dTcat -prefix ./afni_processing/sub_08.results/pb00.sub_08.r01.tcat ds000102/sub-08/func/sub-08_task-flanker_run-1_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.7 s
3dTcat -prefix ./afni_processing/sub_08.results/pb00.sub_08.r02.tcat ds000102/sub-08/func/sub-08_task-flanker_run-2_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.7 s
set tr_counts = ( 146 146 )
cd ./afni_processing/sub_08.results
touch out.pre_ss_warn.txt
foreach run ( 01 02 )
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub_08.r01.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 38915 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r01.1D{0} -expr step(a-0.4)
end
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub_08.r02.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 38896 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r02.1D{0} -expr step(a-0.4)
end
cat outcount.r01.1D outcount.r02.1D
set minindex = `3dTstat -argmin -prefix - outcount_rall.1D\'`
3dTstat -argmin -prefix - outcount_rall.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
set ovals = ( `1d_tool.py -set_run_lengths $tr_counts
-index_to_run_tr $minindex` )
1d_tool.py -set_run_lengths 146 146 -index_to_run_tr 28
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
set minoutrun = 01
set minouttr = 28
echo min outlier: run 01, TR 28
tee out.min_outlier.txt
min outlier: run 01, TR 28
foreach run ( 01 02 )
3dTshift -tzero 0 -quintic -prefix pb01.sub_08.r01.tshift pb00.sub_08.r01.tcat+orig
++ 3dTshift: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: dataset is already aligned in time!
*+ WARNING: ==>> output dataset is just a copy of input dataset
end
3dTshift -tzero 0 -quintic -prefix pb01.sub_08.r02.tshift pb00.sub_08.r02.tcat+orig
++ 3dTshift: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: dataset is already aligned in time!
*+ WARNING: ==>> output dataset is just a copy of input dataset
end
3dbucket -prefix vr_base_min_outlier pb01.sub_08.r01.tshift+orig[28]
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
align_epi_anat.py -anat2epi -anat sub-08_T1w+orig -save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base 0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off
#++ align_epi_anat version: 1.62
#++ turning off volume registration
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./sub-08_T1w+orig
#++ Multi-cost is lpc
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-08_T1w*
#Script is running (command trimmed):
3dcopy ./sub-08_T1w+orig ./__tt_sub-08_T1w+orig
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ Removing skull from anat data
#Script is running (command trimmed):
3dSkullStrip -orig_vol -input ./__tt_sub-08_T1w+orig -prefix ./__tt_sub-08_T1w_ns
#Script is running (command trimmed):
3dinfo ./__tt_sub-08_T1w_ns+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_processing/sub_08.results/__tt_sub-08_T1w_ns+orig is not oblique
#Script is running (command trimmed):
3dinfo ./vr_base_min_outlier+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_processing/sub_08.results/vr_base_min_outlier+orig is not oblique
#++ using 0th sub-brick because only one found
#Script is running (command trimmed):
3dbucket -prefix ./__tt_vr_base_min_outlier_ts ./vr_base_min_outlier+orig'[0]'
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ removing skull or area outside brain
#Script is running (command trimmed):
3dAutomask -apply_prefix ./__tt_vr_base_min_outlier_ts_ns ./__tt_vr_base_min_outlier_ts+orig
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset ./__tt_vr_base_min_outlier_ts+orig
++ Forming automask
+ Fixed clip level = 345.050415
+ Used gradual clip level = 330.013794 .. 365.519012
+ Number voxels above clip level = 39471
+ Clustering voxels ...
+ Largest cluster has 38949 voxels
+ Clustering voxels ...
+ Largest cluster has 38549 voxels
+ Filled 354 voxels in small holes; now have 38903 voxels
+ Filled 3 voxels in large holes; now have 38906 voxels
+ Clustering voxels ...
+ Largest cluster has 38906 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 124934 voxels
+ Mask now has 38906 voxels
++ 38906 voxels in the mask [out of 163840: 23.75%]
++ first 11 x-planes are zero [from L]
++ last 11 x-planes are zero [from R]
++ first 0 y-planes are zero [from P]
++ last 5 y-planes are zero [from A]
++ first 1 z-planes are zero [from I]
++ last 3 z-planes are zero [from S]
++ applying mask to original data
++ Writing masked data
++ Output dataset ./__tt_vr_base_min_outlier_ts_ns+orig.BRIK
++ CPU time = 0.000000 sec
#++ Computing weight mask
#Script is running (command trimmed):
3dBrickStat -automask -percentile 90.000000 1 90.000000 ./__tt_vr_base_min_outlier_ts_ns+orig
#++ Applying threshold of 819.000000 on /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_processing/sub_08.results/__tt_vr_base_min_outlier_ts_ns+orig
#Script is running (command trimmed):
3dcalc -datum float -prefix ./__tt_vr_base_min_outlier_ts_ns_wt -a ./__tt_vr_base_min_outlier_ts_ns+orig -expr 'min(1,(a/819.000000))'
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tt_vr_base_min_outlier_ts_ns_wt+orig.BRIK
#++ Aligning anat data to epi data
#Script is running (command trimmed):
3dAllineate -lpc -wtprefix ./__tt_sub-08_T1w_ns_al_junk_wtal -weight ./__tt_vr_base_min_outlier_ts_ns_wt+orig -source ./__tt_sub-08_T1w_ns+orig -prefix ./sub-08_T1w_al_junk -base ./__tt_vr_base_min_outlier_ts_ns+orig -cmass -1Dmatrix_save ./sub-08_T1w_al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Option '-cmass' enables center-of-mass code = 7 = +xyz
++ Source dataset: ./__tt_sub-08_T1w_ns+orig.HEAD
++ Base dataset: ./__tt_vr_base_min_outlier_ts_ns+orig.HEAD
++ Loading datasets into memory
++ 1583872 voxels in -source_automask+2
++ largeness ==> set -twobest 29
++ Zero-pad: ybot=8 ytop=3
++ Zero-pad: zbot=7 ztop=5
++ 38906 voxels [15.6%] in weight mask
++ Output dataset ./__tt_sub-08_T1w_ns_al_junk_wtal+orig.BRIK
++ Number of points for matching = 38906
++ NOTE: base and source coordinate systems have different handedness
+ Orientations: base=Right handed (LPI); source=Left handed (RPI)
+ - It is nothing to worry about: 3dAllineate aligns based on coordinates.
+ - But it is always important to check the alignment visually to be sure.
++ Local correlation: blok type = 'TOHD(17.0941)'
++ base center of mass = 31.963 35.048 27.741 (index)
+ source center of mass = 84.800 114.693 116.298 (index)
+ source-target CM = 86.190 -3.441 -1.709 (xyz)
+ estimated center of mass shifts = 86.190 -3.441 -1.709
++ shift param auto-range: 25.5..146.9 -74.7..67.8 -67.2..63.8
+ Range param#4 [z-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#5 [x-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#6 [y-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#1 [x-shift] = 76.190132 .. 96.190132 center = 86.190132
+ Range param#2 [y-shift] = -13.440697 .. 6.559303 center = -3.440697
+ Range param#3 [z-shift] = -11.708786 .. 8.291214 center = -1.708786
+ Range param#4 [z-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#5 [x-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#6 [y-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#1 [x-shift] = 46.190132 .. 126.190132 center = 86.190132
+ Range param#2 [y-shift] = -43.440697 .. 36.559303 center = -3.440697
+ Range param#3 [z-shift] = -41.708786 .. 38.291214 center = -1.708786
+ 12 free parameters
++ Normalized (unitless) convergence radius = 0.0000088
++ Final parameter search ranges:
+ x-shift = 46.190 .. 126.190
+ y-shift = -43.441 .. 36.559
+ z-shift = -41.709 .. 38.291
+ z-angle = -45.000 .. 45.000
+ x-angle = -45.000 .. 45.000
+ y-angle = -45.000 .. 45.000
+ x-scale = 0.711 .. 1.406
+ y-scale = 0.711 .. 1.406
+ z-scale = 0.711 .. 1.406
+ y/x-shear = -0.111 .. 0.111
+ z/x-shear = -0.111 .. 0.111
+ z/y-shear = -0.111 .. 0.111
++ changing output grid spacing to 1.0000 mm
++ OpenMP thread count = 15
++ ======= Allineation of 1 sub-bricks using Local Pearson Correlation Signed =======
+ source mask has 1583872 [out of 11534336] voxels
+ base mask has 52253 [out of 249600] voxels
++ ========== sub-brick #0 ========== [total CPU to here=0.0 s]
++ *** Coarse pass begins ***
+ * Enter alignment setup routine
+ - copying base image
+ - copying source image
+ - Smoothing base; radius=4.00
+ - Smoothing source; radius=4.00
+ !source mask fill: ubot=64 usiz=159.5
+ - copying weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ - Search for coarse starting parameters
+ 32391 total points stored in 63 'TOHD(17.5558)' bloks (0 duplicates)
+ - number of free params = 6
+ - Test (64+191)*64 params [top5=*o+-.]:#o+o-.+.o--.--+oo-.++-.o-+-.+o+o---+.--.oo.+++o+$+.--.+..o.o + - best 88 costs found:
o= 0 v=-0.078395: 86.19 -3.44 -1.71 0.00 0.00 0.00 [grid]
o= 1 v=-0.024170: 91.62 4.15 28.38 -38.19 22.74 -41.67 [rand]
o= 2 v=-0.022830: 100.19 -18.97 33.97 -14.61 -39.00 -28.71 [rand]
o= 3 v=-0.022358: 92.86 -30.11 24.96 7.50 -30.00 -30.00 [grid]
o= 4 v=-0.022101: 121.80 -7.85 -38.58 -37.76 24.22 3.86 [rand]
o= 5 v=-0.021851: 97.68 -19.75 -35.46 -19.17 13.96 2.56 [rand]
o= 6 v=-0.021730: 122.84 -20.67 20.55 5.26 -41.39 -40.03 [rand]
o= 7 v=-0.021515: 101.83 -12.20 -35.54 -23.16 17.17 -3.59 [rand]
o= 8 v=-0.021205: 91.12 -0.47 27.39 -40.99 18.21 -33.12 [rand]
o= 9 v=-0.020806: 121.80 -7.85 -38.58 -37.76 24.22 -3.86 [rand]
o=10 v=-0.020703: 97.68 -19.75 -35.46 -19.17 13.96 -2.56 [rand]
o=11 v=-0.020229: 121.80 0.97 35.16 37.76 -24.22 -3.86 [rand]
o=12 v=-0.020075: 90.34 20.37 30.18 31.52 -11.26 -33.63 [rand]
o=13 v=-0.019908: 100.19 -18.97 33.97 14.61 -39.00 -28.71 [rand]
o=14 v=-0.019694: 120.99 -17.22 -35.72 10.62 -7.10 -10.42 [rand]
o=15 v=-0.019540: 119.22 1.58 -38.52 -11.98 4.28 -19.20 [rand]
o=16 v=-0.019170: 88.48 -10.62 -33.40 -10.97 18.37 5.04 [rand]
o=17 v=-0.018913: 121.80 0.97 -38.58 -37.76 24.22 3.86 [rand]
o=18 v=-0.018894: 96.65 -30.71 19.79 -34.95 -5.03 -42.69 [rand]
o=19 v=-0.018891: 115.81 31.17 35.11 24.31 -11.89 -2.84 [rand]
o=20 v=-0.018703: 92.86 -30.11 24.96 -7.50 -30.00 -30.00 [grid]
o=21 v=-0.018636: 115.81 31.17 35.11 24.31 -11.89 2.84 [rand]
o=22 v=-0.018271: 74.70 12.87 -35.46 19.17 13.96 2.56 [rand]
o=23 v=-0.018249: 83.90 -10.62 -33.40 -10.97 18.37 5.04 [rand]
o=24 v=-0.018182: 119.22 -8.46 -38.52 -11.98 4.28 -19.20 [rand]
o=25 v=-0.018157: 112.86 3.23 -28.38 -30.00 7.50 7.50 [grid]
o=26 v=-0.018092: 113.03 -16.54 -34.29 -39.07 28.37 8.01 [rand]
o=27 v=-0.018024: 121.80 -7.85 35.16 37.76 -24.22 -3.86 [rand]
o=28 v=-0.017957: 91.12 -6.41 27.39 -40.99 18.21 -33.12 [rand]
o=29 v=-0.017956: 101.83 -12.20 -35.54 -23.16 17.17 3.59 [rand]
o=30 v=-0.017508: 120.99 -17.22 -35.72 -10.62 -7.10 -10.42 [rand]
o=31 v=-0.017362: 74.70 12.87 -35.46 19.17 13.96 -2.56 [rand]
o=32 v=-0.017349: 120.99 -17.22 -35.72 -10.62 7.10 -10.42 [rand]
o=33 v=-0.016693: 105.31 19.97 -35.01 -37.88 13.88 18.42 [rand]
o=34 v=-0.016675: 102.99 15.02 30.65 10.10 -16.32 -39.31 [rand]
o=35 v=-0.016669: 105.31 19.97 -35.01 -37.88 13.88 -18.42 [rand]
o=36 v=-0.016632: 120.99 -17.22 -35.72 10.62 7.10 -10.42 [rand]
o=37 v=-0.016450: 99.53 -27.69 29.08 6.84 -32.76 -20.90 [rand]
o=38 v=-0.016261: 79.52 -30.11 24.96 -30.00 -7.50 -30.00 [grid]
o=39 v=-0.016134: 99.35 34.44 35.49 14.03 -8.39 -25.20 [rand]
o=40 v=-0.016094: 79.78 30.50 -38.28 -19.28 27.13 15.13 [rand]
o=41 v=-0.015858: 95.39 -15.21 -23.44 -6.82 34.51 24.39 [rand]
o=42 v=-0.015801: 120.99 -17.22 -35.72 10.62 -7.10 10.42 [rand]
o=43 v=-0.015734: 70.55 5.32 -35.54 23.16 17.17 -3.59 [rand]
o=44 v=-0.015723: 67.07 19.97 -35.01 37.88 13.88 -18.42 [rand]
o=45 v=-0.015700: 92.86 23.23 24.96 30.00 -7.50 -30.00 [grid]
o=46 v=-0.015623: 113.12 -31.58 -39.65 15.05 4.71 -12.78 [rand]
o=47 v=-0.015567: 92.86 -10.11 -28.38 -7.50 7.50 30.00 [grid]
o=48 v=-0.015330: 88.48 -10.62 -33.40 -10.97 18.37 -5.04 [rand]
o=49 v=-0.015192: 92.03 -32.53 20.97 30.43 -41.57 -36.66 [rand]
o=50 v=-0.015111: 103.24 -26.13 22.54 -12.15 -12.44 -39.75 [rand]
o=51 v=-0.015075: 122.78 -30.74 29.53 -25.97 -37.53 -40.21 [rand]
o=52 v=-0.014925: 92.86 -10.11 -28.38 -7.50 30.00 30.00 [grid]
o=53 v=-0.014904: 121.80 0.97 -38.58 -37.76 24.22 -3.86 [rand]
o=54 v=-0.014835: 102.99 -21.90 30.65 -10.10 -16.32 -39.31 [rand]
o=55 v=-0.014711: 102.99 15.02 -34.07 -10.10 -16.32 39.31 [rand]
o=56 v=-0.014651: 119.22 -8.46 -38.52 -11.98 -4.28 -19.20 [rand]
o=57 v=-0.014527: 99.35 -41.33 -38.90 14.03 8.39 -25.20 [rand]
o=58 v=-0.014469: 70.55 -12.20 -35.54 23.16 17.17 3.59 [rand]
o=59 v=-0.014214: 121.80 -7.85 35.16 37.76 -24.22 3.86 [rand]
o=60 v=-0.014171: 70.55 -12.20 -35.54 23.16 17.17 -3.59 [rand]
o=61 v=-0.014132: 94.85 -27.60 34.86 27.41 -42.10 -24.35 [rand]
o=62 v=-0.014108: 92.86 -10.11 -28.38 30.00 7.50 7.50 [grid]
o=63 v=-0.013985: 92.61 -37.38 -38.28 19.28 27.13 -15.13 [rand]
o=64 v=-0.013771: 56.91 -19.20 28.15 13.45 -19.96 14.96 [rand]
o=65 v=-0.013738: 104.93 -39.84 -34.76 24.90 4.74 -17.23 [rand]
o=66 v=-0.013600: 92.86 3.23 -28.38 -7.50 30.00 30.00 [grid]
o=67 v=-0.013545: 102.99 15.02 -34.07 10.10 -16.32 39.31 [rand]
o=68 v=-0.013506: 97.68 12.87 -35.46 -19.17 13.96 -2.56 [rand]
o=69 v=-0.013420: 92.86 3.23 24.96 30.00 -30.00 -30.00 [grid]
o=70 v=-0.013384: 83.90 -10.62 -33.40 -10.97 18.37 -5.04 [rand]
o=71 v=-0.013341: 115.81 -38.05 -38.52 24.31 -11.89 -2.84 [rand]
o=72 v=-0.013295: 59.26 -31.58 36.24 15.05 -4.71 12.78 [rand]
o=73 v=-0.013221: 79.78 30.50 -38.28 19.28 27.13 15.13 [rand]
o=74 v=-0.013191: 59.35 9.66 -34.29 39.07 28.37 -8.01 [rand]
o=75 v=-0.013149: 118.40 -32.49 34.43 -40.41 -19.09 -40.75 [rand]
o=76 v=-0.013072: 70.55 5.32 -35.54 23.16 17.17 3.59 [rand]
o=77 v=-0.013043: 65.94 23.84 -36.19 37.90 5.03 3.08 [rand]
o=78 v=-0.012929: 92.86 -10.11 -28.38 7.50 7.50 7.50 [grid]
o=79 v=-0.012857: 60.67 -34.78 32.57 42.46 -41.07 35.63 [rand]
o=80 v=-0.012789: 99.35 -41.33 35.49 14.03 -8.39 -25.20 [rand]
o=81 v=-0.012665: 97.68 12.87 -35.46 -19.17 13.96 2.56 [rand]
o=82 v=-0.012652: 112.86 -30.11 -28.38 30.00 7.50 7.50 [grid]
o=83 v=-0.012596: 104.93 -39.84 31.34 -24.90 -4.74 -17.23 [rand]
o=84 v=-0.012570: 83.90 -10.62 -33.40 10.97 18.37 -5.04 [rand]
o=85 v=-0.012553: 92.86 -10.11 -28.38 -7.50 30.00 7.50 [grid]
o=86 v=-0.012529: 92.86 -10.11 24.96 -7.50 -7.50 -30.00 [grid]
o=87 v=-0.012519: 80.37 10.75 27.93 38.21 -15.41 -13.51 [rand]
+ - A little optimization:*[#16335=-0.0833147] *[#16338=-0.0996876] *[#16339=-0.122258] *[#16342=-0.125257] *[#16344=-0.126031] *[#16346=-0.127847] *[#16349=-0.129014] *[#16352=-0.129155] *[#16355=-0.129328] *[#16356=-0.129367] *[#16360=-0.12939] *[#16362=-0.129425] *[#16363=-0.129437] *[#16366=-0.129478] ........................................................................................
+ - costs of the above after a little optimization:
*o= 0 v=-0.129478: 86.56 -2.92 0.24 -1.17 0.39 0.22 [grid] [f=50]
o= 1 v=-0.025208: 91.85 3.62 28.37 -37.98 22.65 -41.84 [rand] [f=41]
o= 2 v=-0.037638: 97.07 -22.70 36.86 -10.25 -41.88 -25.72 [rand] [f=62]
o= 3 v=-0.026228: 94.01 -30.11 26.00 6.89 -35.26 -29.95 [grid] [f=67]
o= 4 v=-0.025109: 122.87 -3.88 -38.32 -37.64 24.31 3.06 [rand] [f=61]
o= 5 v=-0.023467: 97.59 -19.20 -35.11 -18.55 14.45 2.78 [rand] [f=47]
o= 6 v=-0.032558: 124.34 -28.56 20.34 5.63 -41.94 -44.51 [rand] [f=52]
o= 7 v=-0.024558: 103.87 -12.91 -36.21 -26.12 14.64 -5.56 [rand] [f=68]
o= 8 v=-0.026595: 92.49 0.17 27.38 -37.57 16.88 -34.60 [rand] [f=47]
o= 9 v=-0.025748: 122.87 -7.62 -36.96 -38.73 25.59 5.74 [rand] [f=37]
o=10 v=-0.023050: 97.81 -19.65 -34.79 -19.15 13.89 2.05 [rand] [f=36]
o=11 v=-0.034817: 125.56 3.51 37.77 32.42 -27.27 -8.86 [rand] [f=47]
o=12 v=-0.026636: 90.62 19.43 30.46 30.75 -11.47 -27.04 [rand] [f=54]
o=13 v=-0.029615: 95.65 -16.42 33.99 15.06 -38.94 -27.36 [rand] [f=44]
o=14 v=-0.025724: 121.05 -17.94 -34.73 10.44 -8.00 -4.28 [rand] [f=40]
o=15 v=-0.027042: 119.10 0.10 -39.89 -13.81 11.33 -14.01 [rand] [f=42]
o=16 v=-0.029803: 88.38 -15.30 -33.36 -10.38 12.73 4.37 [rand] [f=58]
o=17 v=-0.025471: 120.77 -7.35 -37.30 -34.55 25.97 6.34 [rand] [f=51]
o=18 v=-0.021813: 96.06 -31.35 20.30 -37.16 -7.69 -44.56 [rand] [f=37]
o=19 v=-0.039610: 120.38 31.62 37.96 31.21 -11.89 -1.49 [rand] [f=60]
o=20 v=-0.030208: 99.98 -30.12 26.92 -4.50 -28.25 -34.17 [grid] [f=62]
o=21 v=-0.033973: 115.95 30.51 37.09 27.08 -7.23 4.45 [rand] [f=45]
o=22 v=-0.024448: 68.66 12.44 -38.44 19.11 14.37 2.63 [rand] [f=52]
o=23 v=-0.030816: 88.36 -12.05 -34.28 -12.31 11.56 5.36 [rand] [f=57]
o=24 v=-0.026831: 119.70 -7.09 -37.52 -10.99 4.50 -13.47 [rand] [f=43]
o=25 v=-0.021834: 110.54 4.11 -28.89 -31.19 6.72 7.75 [grid] [f=46]
o=26 v=-0.029659: 112.69 -13.96 -35.31 -28.09 23.99 4.39 [rand] [f=64]
o=27 v=-0.036380: 124.25 -11.62 38.17 33.28 -31.14 -4.50 [rand] [f=57]
o=28 v=-0.026330: 92.18 1.00 27.77 -39.33 18.23 -32.61 [rand] [f=51]
o=29 v=-0.022613: 102.63 -11.90 -37.10 -25.08 13.86 -0.27 [rand] [f=35]
o=30 v=-0.023502: 119.40 -16.34 -38.41 -9.52 -2.70 -13.24 [rand] [f=49]
o=31 v=-0.024601: 69.46 14.02 -37.34 19.44 12.69 -2.24 [rand] [f=44]
o=32 v=-0.025824: 119.43 -8.11 -37.98 -7.01 10.78 -13.52 [rand] [f=83]
o=33 v=-0.028485: 111.32 17.78 -33.51 -44.98 10.27 21.60 [rand] [f=50]
o=34 v=-0.021606: 103.05 11.34 30.91 10.44 -16.63 -39.34 [rand] [f=54]
o=35 v=-0.025812: 105.92 18.80 -34.03 -38.48 13.27 -12.88 [rand] [f=49]
o=36 v=-0.023639: 120.56 -21.02 -38.02 5.46 4.63 -15.69 [rand] [f=51]
o=37 v=-0.022421: 96.59 -28.00 29.09 6.71 -31.63 -25.55 [rand] [f=39]
o=38 v=-0.020190: 79.59 -26.31 24.88 -29.92 -8.68 -28.78 [grid] [f=49]
o=39 v=-0.021749: 96.94 33.57 35.86 11.35 -6.94 -19.87 [rand] [f=66]
o=40 v=-0.019895: 82.95 28.96 -37.93 -17.75 28.76 13.02 [rand] [f=37]
o=41 v=-0.021356: 91.66 -14.96 -22.76 -7.10 34.58 24.82 [rand] [f=46]
o=42 v=-0.027107: 124.49 -20.18 -32.05 10.69 -7.44 9.91 [rand] [f=45]
o=43 v=-0.026319: 68.91 11.17 -38.05 20.29 19.40 -6.43 [rand] [f=56]
o=44 v=-0.024913: 67.84 15.42 -35.49 37.39 14.37 -18.96 [rand] [f=37]
o=45 v=-0.023354: 93.76 23.30 27.77 29.47 -7.75 -31.27 [grid] [f=40]
o=46 v=-0.019407: 109.48 -31.82 -39.14 15.51 4.62 -13.43 [rand] [f=46]
o=47 v=-0.022879: 92.79 -14.50 -30.57 -6.67 8.67 35.86 [grid] [f=59]
o=48 v=-0.028698: 88.61 -13.59 -36.48 -10.26 12.62 0.84 [rand] [f=53]
o=49 v=-0.019705: 92.37 -28.32 21.44 30.84 -39.72 -37.84 [rand] [f=65]
o=50 v=-0.022741: 99.40 -24.95 21.80 -13.00 -13.24 -40.83 [rand] [f=46]
o=51 v=-0.025023: 125.97 -26.96 37.93 -27.17 -36.78 -38.74 [rand] [f=57]
o=52 v=-0.018040: 92.91 -9.23 -27.61 -8.65 30.90 29.96 [grid] [f=65]
o=53 v=-0.026083: 119.13 -8.06 -39.09 -31.95 24.41 -0.92 [rand] [f=47]
o=54 v=-0.021481: 97.21 -15.76 28.19 -9.68 -14.20 -39.20 [rand] [f=52]
o=55 v=-0.029622: 102.22 23.54 -40.40 -8.51 -15.66 40.94 [rand] [f=57]
o=56 v=-0.024318: 119.26 -12.01 -40.99 -6.33 -3.92 -15.68 [rand] [f=67]
o=57 v=-0.016309: 99.90 -40.50 -41.08 13.83 7.03 -28.14 [rand] [f=41]
o=58 v=-0.020067: 70.82 -7.68 -36.96 23.60 18.14 3.28 [rand] [f=53]
o=59 v=-0.029083: 122.02 -8.54 38.29 35.74 -24.75 2.19 [rand] [f=40]
o=60 v=-0.018760: 69.81 -16.27 -37.23 22.97 16.08 -3.59 [rand] [f=33]
o=61 v=-0.027637: 95.70 -14.89 33.59 24.68 -37.62 -25.78 [rand] [f=55]
o=62 v=-0.021869: 90.87 -7.14 -30.34 27.19 5.03 7.77 [grid] [f=44]
o=63 v=-0.016562: 93.04 -33.28 -38.26 19.24 27.71 -15.44 [rand] [f=35]
o=64 v=-0.017593: 53.52 -19.20 30.34 13.28 -18.83 16.03 [rand] [f=60]
o=65 v=-0.020603: 105.13 -39.25 -35.37 22.69 13.32 -21.57 [rand] [f=50]
o=66 v=-0.022279: 93.18 9.76 -30.87 -8.28 32.53 27.85 [grid] [f=76]
o=67 v=-0.023893: 102.80 20.21 -36.92 8.62 -14.85 40.53 [rand] [f=50]
o=68 v=-0.030230: 94.78 11.44 -41.70 -27.01 18.02 -15.08 [rand] [f=53]
o=69 v=-0.025604: 92.34 2.49 30.25 29.16 -28.24 -27.67 [grid] [f=63]
o=70 v=-0.028099: 84.44 -14.51 -37.03 -5.85 10.74 5.83 [rand] [f=62]
o=71 v=-0.024499: 117.76 -30.97 -33.13 30.69 -12.85 -3.72 [rand] [f=61]
o=72 v=-0.017104: 59.48 -35.24 36.02 15.19 -4.00 12.18 [rand] [f=44]
o=73 v=-0.020121: 78.49 34.05 -38.37 19.48 26.09 14.47 [rand] [f=43]
o=74 v=-0.027725: 66.37 14.95 -34.10 33.91 20.53 -10.37 [rand] [f=70]
o=75 v=-0.021457: 118.15 -34.24 32.27 -35.98 -26.19 -42.12 [rand] [f=53]
o=76 v=-0.033583: 84.74 5.08 -40.21 22.43 10.74 3.93 [rand] [f=82]
o=77 v=-0.022471: 66.16 19.67 -38.88 38.58 4.87 -0.04 [rand] [f=62]
o=78 v=-0.022165: 93.72 -7.71 -26.28 17.15 7.89 8.13 [grid] [f=39]
o=79 v=-0.019392: 56.80 -35.73 32.75 42.93 -41.01 35.05 [rand] [f=40]
o=80 v=-0.024286: 99.09 -41.34 36.23 14.84 -11.57 -20.31 [rand] [f=40]
o=81 v=-0.027989: 100.32 13.71 -36.07 -26.64 6.16 -4.67 [rand] [f=61]
o=82 v=-0.018578: 114.04 -25.07 -27.78 31.22 5.83 3.98 [grid] [f=41]
o=83 v=-0.020005: 108.98 -40.26 31.30 -22.78 -4.31 -16.42 [rand] [f=52]
o=84 v=-0.022922: 85.64 -10.65 -38.37 5.12 14.67 -7.14 [rand] [f=60]
o=85 v=-0.022479: 89.13 -11.37 -28.03 -5.87 37.76 7.84 [grid] [f=59]
o=86 v=-0.019822: 93.58 -9.73 28.66 -9.65 -11.27 -31.86 [grid] [f=57]
o=87 v=-0.031659: 80.56 10.85 31.85 39.61 -23.66 -12.54 [rand] [f=68]
+ - saving # 0 for use with twobest
+ - saving #19 for use with twobest
+ - saving # 2 for use with twobest
+ - saving #27 for use with twobest
+ - saving #11 for use with twobest
+ - saving #21 for use with twobest
+ - saving #76 for use with twobest
+ - saving # 6 for use with twobest
+ - saving #87 for use with twobest
+ - saving #23 for use with twobest
+ - saving #68 for use with twobest
+ - saving #20 for use with twobest
+ - skip #16 for twobest: too close to set #23
+ - saving #26 for use with twobest
+ - saving #55 for use with twobest
+ - saving #13 for use with twobest
+ - saving #59 for use with twobest
+ - skip #48 for twobest: too close to set #16
+ - saving #33 for use with twobest
+ - saving #70 for use with twobest
+ - saving #81 for use with twobest
+ - saving #74 for use with twobest
+ - saving #61 for use with twobest
+ - saving #42 for use with twobest
+ - saving #15 for use with twobest
+ - saving #24 for use with twobest
+ - saving #12 for use with twobest
+ - saving # 8 for use with twobest
+ - skip #28 for twobest: too close to set # 8
+ - saving #43 for use with twobest
+ - saving # 3 for use with twobest
+ - saving #53 for use with twobest
+ - Coarse startup search net CPU time = 0.0 s
++ Start refinement #1 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=3.11
+ - Smoothing source; radius=3.11
+ !source mask fill: ubot=64 usiz=159.5
+ - retaining old weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31909 total points stored in 63 'TOHD(17.3748)' bloks (0 duplicates)
*[#20978=-0.169331] *[#20986=-0.170313] *[#21000=-0.170526] *[#21001=-0.172023] *[#21005=-0.172274] *[#21008=-0.173681] *[#21009=-0.1739] *[#21013=-0.174339] *[#21016=-0.175108] *[#21017=-0.176175] *[#21018=-0.176452] *[#21019=-0.176809] *[#21021=-0.177261] *[#21024=-0.17761] *[#21026=-0.178186] *[#21027=-0.178833] *[#21028=-0.179343] *[#21031=-0.179472] *[#21040=-0.180395] *[#21043=-0.180428] *[#21045=-0.180429] *[#21047=-0.180514] *[#21048=-0.180599] *[#21049=-0.180672] *[#21051=-0.180752] *[#21052=-0.180786] *[#21053=-0.180808] *[#21054=-0.18082] *[#21055=-0.180832] *[#21056=-0.180858] *[#21057=-0.180864] *[#21059=-0.180909] *[#21060=-0.180915] *[#21061=-0.180934] *[#21062=-0.181065] *[#21063=-0.181087] *[#21064=-0.181114] *[#21065=-0.181253] *[#21068=-0.181324] *[#21069=-0.181432] *[#21070=-0.18147] *[#21073=-0.181504] *[#21074=-0.18155] *[#21075=-0.181565]
+ - param set #1 has cost=-0.181565 [o=0 t=0]
+ -- Parameters = 86.2417 -3.0340 0.1839 -0.7927 0.0198 0.1572 0.9849 1.0186 0.9937 -0.0067 -0.0009 -0.0030
+ - param set #2 has cost=-0.029599 [o=19 t=1]
+ -- Parameters = 120.3747 31.4269 37.8606 31.0260 -11.8449 -1.1094 1.0067 0.9979 0.9877 -0.0009 0.0021 -0.0001
+ - param set #3 has cost=-0.032947 [o=2 t=2]
+ -- Parameters = 97.1422 -22.9068 36.7125 -10.4271 -42.0244 -25.8414 0.9953 0.9994 0.9958 -0.0016 0.0013 -0.0002
+ - param set #4 has cost=-0.025913 [o=27 t=3]
+ -- Parameters = 124.2213 -11.7386 37.9231 33.3757 -31.0928 -4.5253 1.0015 1.0012 0.9970 0.0002 0.0015 0.0004
+ - param set #5 has cost=-0.028112 [o=11 t=4]
+ -- Parameters = 122.3987 3.2658 37.8554 32.0364 -27.2812 -8.8197 0.9993 1.0000 0.9989 -0.0002 -0.0001 -0.0002
+ - param set #6 has cost=-0.027098 [o=21 t=5]
+ -- Parameters = 115.6655 30.6513 37.9152 27.7891 -10.1989 1.4674 0.9984 0.9912 0.9884 0.0004 -0.0012 0.0008
+ - param set #7 has cost=-0.025330 [o=76 t=6]
+ -- Parameters = 84.5296 4.8670 -40.1157 22.4018 10.4986 3.9660 1.0038 0.9999 1.0018 -0.0005 -0.0003 0.0016
+ - param set #8 has cost=-0.027671 [o=6 t=7]
+ -- Parameters = 124.4184 -28.3909 20.3705 5.2792 -41.8702 -44.1177 1.0047 0.9790 1.0062 -0.0035 -0.0002 -0.0054
+ - param set #9 has cost=-0.026880 [o=87 t=8]
+ -- Parameters = 80.6211 10.7956 31.8627 39.3826 -23.6462 -12.7049 0.9977 1.0051 0.9899 -0.0002 0.0006 -0.0006
+ - param set #10 has cost=-0.025438 [o=23 t=9]
+ -- Parameters = 88.5119 -12.3057 -34.5255 -12.2625 11.3388 4.8335 1.0036 0.9951 0.9911 0.0147 -0.0003 -0.0010
+ - param set #11 has cost=-0.025056 [o=68 t=10]
+ -- Parameters = 95.0321 11.2848 -41.6982 -27.1384 18.0539 -14.7625 0.9967 0.9975 1.0046 0.0010 -0.0017 -0.0002
+ - param set #12 has cost=-0.026301 [o=20 t=11]
+ -- Parameters = 99.7295 -29.7606 26.8107 -4.8322 -28.5727 -34.7626 1.0025 0.9939 1.0000 0.0001 -0.0010 0.0011
+ - param set #13 has cost=-0.024785 [o=26 t=12]
+ -- Parameters = 112.7484 -13.9672 -35.1899 -28.0512 24.1536 4.0959 0.9974 1.0021 1.0031 -0.0009 0.0004 -0.0004
+ - param set #14 has cost=-0.023818 [o=55 t=13]
+ -- Parameters = 102.3059 23.5118 -40.3609 -8.3749 -15.9182 41.1731 0.9987 1.0004 1.0059 0.0001 -0.0012 -0.0005
+ - param set #15 has cost=-0.025346 [o=13 t=14]
+ -- Parameters = 95.5049 -16.5664 34.3042 14.7561 -38.9196 -27.3528 1.0042 1.0061 0.9993 0.0005 -0.0002 -0.0014
+ - param set #16 has cost=-0.028890 [o=59 t=15]
+ -- Parameters = 121.0495 -8.3479 38.2577 35.4740 -28.4600 -2.1114 0.9475 0.9673 0.9742 0.0001 -0.0099 0.0125
+ - param set #17 has cost=-0.022857 [o=33 t=16]
+ -- Parameters = 111.2125 17.7604 -33.8452 -44.8026 10.9680 21.5318 0.9972 1.0001 1.0062 -0.0000 0.0110 -0.0001
+ - param set #18 has cost=-0.024121 [o=70 t=17]
+ -- Parameters = 84.6139 -14.8654 -35.5807 -6.2385 9.4374 4.5597 0.9971 1.0019 0.9957 -0.0008 0.0001 -0.0020
+ - param set #19 has cost=-0.026862 [o=81 t=18]
+ -- Parameters = 100.4019 15.0228 -35.6483 -31.5537 6.5158 -4.5506 0.9870 0.9952 0.9971 0.0007 0.0006 0.0008
+ - param set #20 has cost=-0.019718 [o=74 t=19]
+ -- Parameters = 66.3511 14.9204 -34.0041 33.9302 20.6377 -10.4785 0.9995 1.0003 0.9990 0.0002 -0.0001 -0.0002
+ - param set #21 has cost=-0.024596 [o=61 t=20]
+ -- Parameters = 94.9530 -15.7971 34.3648 21.1914 -38.2038 -26.5993 0.9967 1.0153 0.9925 0.0030 0.0004 -0.0029
+ - param set #22 has cost=-0.030211 [o=42 t=21]
+ -- Parameters = 125.4314 -20.5678 -30.0616 11.1232 -9.1532 7.8234 0.9842 0.9823 0.9478 -0.0144 -0.0209 0.0117
+ - param set #23 has cost=-0.022798 [o=15 t=22]
+ -- Parameters = 119.2795 0.0863 -39.9379 -13.8465 11.2937 -14.2121 0.9972 0.9982 1.0023 0.0015 -0.0005 -0.0005
+ - param set #24 has cost=-0.022617 [o=24 t=23]
+ -- Parameters = 119.5177 -7.0698 -37.4677 -10.8037 4.7487 -13.3916 1.0004 1.0044 0.9954 0.0003 -0.0039 -0.0012
+ - param set #25 has cost=-0.025054 [o=12 t=24]
+ -- Parameters = 90.2415 19.2854 31.2812 30.8214 -12.2280 -26.6292 0.9929 0.9856 0.9532 -0.0054 0.0058 -0.0182
+ - param set #26 has cost=-0.022589 [o=8 t=25]
+ -- Parameters = 92.2565 -0.3541 27.7222 -37.8739 16.7082 -34.1457 1.0015 0.9972 0.9965 0.0001 0.0001 -0.0009
+ - param set #27 has cost=-0.023658 [o=43 t=26]
+ -- Parameters = 67.8001 10.6026 -38.1869 20.2024 17.6918 -6.5006 0.9989 0.9962 0.9715 0.0005 -0.0034 0.0218
+ - param set #28 has cost=-0.023512 [o=3 t=27]
+ -- Parameters = 93.6271 -30.3348 25.2672 6.4093 -34.2989 -30.7456 1.0028 0.9979 1.0102 0.0018 0.0004 -0.0028
+ - param set #29 has cost=-0.022179 [o=53 t=28]
+ -- Parameters = 119.1053 -8.1075 -39.0073 -32.2735 24.1427 -0.5844 0.9994 1.0016 1.0011 -0.0012 0.0000 0.0001
*[#23803=-0.181994] *[#23804=-0.182059] *[#23808=-0.182443] *[#23813=-0.182802] *[#23817=-0.182824] *[#23821=-0.183125]
+ - param set #30 has cost=-0.183125 [o=-1 t=-1]
+ -- Parameters = 86.2790 -2.9386 0.2322 -0.5467 -0.0139 0.1276 0.9790 1.0183 0.9889 0.0075 0.0058 -0.0075
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0636
+ --- dist(#3,#1) = 0.467
+ --- dist(#4,#1) = 0.489
+ --- dist(#5,#1) = 0.47
+ --- dist(#6,#1) = 0.475
+ --- dist(#7,#1) = 0.47
+ --- dist(#8,#1) = 0.492
+ --- dist(#9,#1) = 0.471
+ --- dist(#10,#1) = 0.444
+ --- dist(#11,#1) = 0.449
+ --- dist(#12,#1) = 0.388
+ --- dist(#13,#1) = 0.474
+ --- dist(#14,#1) = 0.434
+ --- dist(#15,#1) = 0.432
+ --- dist(#16,#1) = 0.504
+ --- dist(#17,#1) = 0.524
+ --- dist(#18,#1) = 0.388
+ --- dist(#19,#1) = 0.443
+ --- dist(#20,#1) = 0.427
+ --- dist(#21,#1) = 0.448
+ --- dist(#22,#1) = 0.507
+ --- dist(#23,#1) = 0.48
+ --- dist(#24,#1) = 0.381
+ --- dist(#25,#1) = 0.492
+ --- dist(#26,#1) = 0.502
+ --- dist(#27,#1) = 0.471
+ --- dist(#28,#1) = 0.415
+ --- dist(#29,#1) = 0.49
+ --- dist(#30,#1) = 0.428
++ Start refinement #2 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=2.42
+ - Smoothing source; radius=2.42
+ !source mask fill: ubot=64 usiz=159.5
+ - retaining old weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31999 total points stored in 64 'TOHD(17.2644)' bloks (0 duplicates)
*[#23823=-0.208583] *[#23853=-0.209083] *[#23856=-0.209692] *[#23857=-0.21004] *[#23858=-0.210473] *[#23859=-0.210652] *[#23861=-0.210912] *[#23862=-0.21108] *[#23863=-0.211096] *[#23864=-0.211318] *[#23865=-0.211465] *[#23868=-0.211558] *[#23871=-0.211598] *[#23874=-0.211605] *[#23876=-0.211804] *[#23880=-0.211892] *[#23883=-0.212487] *[#23884=-0.212638] *[#23887=-0.212807] *[#23889=-0.212858] *[#23890=-0.212946] *[#23891=-0.213045] *[#23892=-0.213054] *[#23893=-0.213085] *[#23896=-0.213618] *[#23897=-0.214048] *[#23898=-0.2143] *[#23899=-0.214697] *[#23902=-0.214782] *[#23904=-0.215131] *[#23907=-0.215508] *[#23908=-0.21602] *[#23909=-0.21655] *[#23910=-0.216938] *[#23911=-0.21729] *[#23912=-0.217316] *[#23913=-0.217499] *[#23914=-0.217762] *[#23915=-0.217858] *[#23920=-0.218401] *[#23921=-0.218462] *[#23922=-0.218657] *[#23925=-0.219463] *[#23926=-0.219544] *[#23927=-0.219959] *[#23928=-0.220151] *[#23931=-0.220227]
+ - param set #1 has cost=-0.220227 [o=-1 t=-1]
+ -- Parameters = 86.5690 -3.1646 -0.1381 -0.8542 -1.9133 -0.7763 0.9848 1.0171 0.9535 -0.0108 0.0209 -0.0382
+ - param set #2 has cost=-0.216445 [o=0 t=0]
+ -- Parameters = 86.4148 -3.1639 -0.1179 -0.8652 -0.7590 -0.0405 0.9867 1.0242 0.9680 -0.0151 0.0021 -0.0159
+ - param set #3 has cost=-0.027810 [o=2 t=2]
+ -- Parameters = 97.2032 -22.9052 36.6921 -10.4554 -42.1013 -25.7685 0.9914 0.9972 0.9931 -0.0021 0.0004 0.0019
+ - param set #4 has cost=-0.030534 [o=42 t=21]
+ -- Parameters = 124.9320 -22.0031 -26.3056 8.7231 -2.9528 10.9929 0.9078 0.9429 0.9127 -0.0312 -0.0436 -0.0116
+ - param set #5 has cost=-0.024661 [o=19 t=1]
+ -- Parameters = 120.2512 31.5807 37.8833 31.4755 -11.7813 -0.9877 1.0280 0.9922 0.9858 -0.0015 0.0013 0.0011
+ - param set #6 has cost=-0.022586 [o=59 t=15]
+ -- Parameters = 121.0138 -8.5765 38.2813 35.5698 -28.5095 -2.1222 0.9444 0.9668 0.9787 0.0009 -0.0099 0.0127
+ - param set #7 has cost=-0.024438 [o=11 t=4]
+ -- Parameters = 122.3828 3.2625 37.8905 32.0936 -27.2969 -8.9957 0.9996 1.0008 0.9996 -0.0003 -0.0004 0.0002
+ - param set #8 has cost=-0.022973 [o=6 t=7]
+ -- Parameters = 124.5990 -28.5221 20.3574 5.1992 -41.4910 -44.5459 1.0133 0.9665 0.9986 -0.0044 -0.0021 -0.0082
+ - param set #9 has cost=-0.023141 [o=21 t=5]
+ -- Parameters = 115.3082 31.2509 37.9653 28.2268 -10.2700 2.1299 0.9881 0.9725 1.0137 0.0031 -0.0002 -0.0013
+ - param set #10 has cost=-0.020712 [o=87 t=8]
+ -- Parameters = 80.6871 10.7820 31.8608 39.2432 -23.8256 -12.8107 0.9960 1.0010 0.9862 0.0001 -0.0012 -0.0004
+ - param set #11 has cost=-0.023867 [o=81 t=18]
+ -- Parameters = 100.9144 15.5630 -35.5687 -30.9887 7.2817 -4.8546 0.9920 1.0234 0.9953 -0.0013 0.0007 -0.0033
+ - param set #12 has cost=-0.022773 [o=20 t=11]
+ -- Parameters = 99.3746 -29.8610 26.9858 -5.3618 -28.5046 -34.3145 1.0066 0.9859 1.0136 -0.0014 0.0000 0.0010
+ - param set #13 has cost=-0.021875 [o=27 t=3]
+ -- Parameters = 124.5604 -15.0908 37.9811 33.1165 -31.3178 -4.2711 1.0069 1.0029 0.9948 0.0004 0.0007 0.0018
+ - param set #14 has cost=-0.018972 [o=23 t=9]
+ -- Parameters = 88.7944 -12.1116 -34.5232 -12.0411 11.5798 4.8169 1.0017 0.9955 0.9899 0.0150 0.0007 -0.0000
+ - param set #15 has cost=-0.022093 [o=13 t=14]
+ -- Parameters = 94.6722 -16.0639 34.9181 15.7936 -38.3548 -28.1825 0.9908 0.9977 0.9904 -0.0019 -0.0004 -0.0028
+ - param set #16 has cost=-0.019604 [o=76 t=6]
+ -- Parameters = 84.2874 4.5998 -40.1267 21.8844 10.7009 4.2715 1.0055 0.9996 1.0026 -0.0011 -0.0010 0.0040
+ - param set #17 has cost=-0.018545 [o=68 t=10]
+ -- Parameters = 94.9844 11.4663 -41.6971 -27.1688 18.1245 -14.7945 0.9941 1.0035 1.0045 0.0090 -0.0015 -0.0008
+ - param set #18 has cost=-0.022035 [o=12 t=24]
+ -- Parameters = 90.4553 19.1335 31.0461 30.9460 -12.3561 -27.0333 0.9931 0.9841 0.9491 -0.0063 0.0070 -0.0183
+ - param set #19 has cost=-0.017809 [o=26 t=12]
+ -- Parameters = 112.7930 -14.0675 -34.9825 -28.0190 24.1066 3.9824 0.9955 1.0005 1.0055 -0.0008 0.0082 -0.0007
+ - param set #20 has cost=-0.022233 [o=61 t=20]
+ -- Parameters = 94.9835 -15.7922 34.2528 21.3591 -38.3023 -26.6743 0.9942 1.0155 0.9949 0.0025 0.0007 -0.0032
+ - param set #21 has cost=-0.021028 [o=70 t=17]
+ -- Parameters = 86.4718 -14.4547 -34.6323 -6.2583 9.6649 3.7010 0.9942 0.9844 0.9884 0.0090 0.0005 -0.0028
+ - param set #22 has cost=-0.017872 [o=55 t=13]
+ -- Parameters = 102.4708 23.5267 -40.2705 -8.5359 -16.0124 41.0981 1.0009 0.9972 1.0084 -0.0001 -0.0017 -0.0010
+ - param set #23 has cost=-0.017721 [o=43 t=26]
+ -- Parameters = 67.4275 10.6734 -38.2748 20.0915 17.9131 -6.3259 0.9971 0.9935 0.9725 0.0008 0.0052 0.0220
+ - param set #24 has cost=-0.020349 [o=3 t=27]
+ -- Parameters = 92.5580 -30.5120 25.0446 5.9667 -33.9224 -32.3783 0.9901 0.9827 0.9962 0.0134 0.0054 -0.0044
+ - param set #25 has cost=-0.018272 [o=33 t=16]
+ -- Parameters = 110.7879 17.5217 -34.1652 -44.9186 11.3923 22.6426 0.9815 0.9941 0.9763 -0.0011 0.0224 -0.0024
+ - param set #26 has cost=-0.016165 [o=15 t=22]
+ -- Parameters = 119.3371 0.1366 -40.0015 -13.9032 11.2916 -14.2405 0.9960 0.9969 1.0033 0.0023 -0.0007 -0.0005
+ - param set #27 has cost=-0.017790 [o=24 t=23]
+ -- Parameters = 119.5484 -7.0770 -37.5706 -10.9167 4.8044 -13.1528 1.0001 1.0034 0.9886 -0.0026 0.0023 -0.0026
+ - param set #28 has cost=-0.021054 [o=8 t=25]
+ -- Parameters = 92.1249 -0.7941 28.1167 -37.9315 16.4735 -34.0354 0.9818 0.9814 0.9791 -0.0018 -0.0013 -0.0009
+ - param set #29 has cost=-0.015293 [o=53 t=28]
+ -- Parameters = 119.0922 -8.1040 -38.9836 -32.2314 24.1018 -0.5725 1.0000 1.0027 1.0031 -0.0012 0.0001 0.0002
+ - param set #30 has cost=-0.014662 [o=74 t=19]
+ -- Parameters = 66.4003 14.9545 -34.0002 33.9259 20.6687 -10.5010 0.9998 1.0006 0.9992 0.0001 -0.0000 0.0007
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.1
+ --- dist(#3,#1) = 0.48
+ --- dist(#4,#1) = 0.46
+ --- dist(#5,#1) = 0.475
+ --- dist(#6,#1) = 0.475
+ --- dist(#7,#1) = 0.443
+ --- dist(#8,#1) = 0.476
+ --- dist(#9,#1) = 0.486
+ --- dist(#10,#1) = 0.373
+ --- dist(#11,#1) = 0.48
+ --- dist(#12,#1) = 0.43
+ --- dist(#13,#1) = 0.438
+ --- dist(#14,#1) = 0.39
+ --- dist(#15,#1) = 0.476
+ --- dist(#16,#1) = 0.412
+ --- dist(#17,#1) = 0.431
+ --- dist(#18,#1) = 0.446
+ --- dist(#19,#1) = 0.356
+ --- dist(#20,#1) = 0.5
+ --- dist(#21,#1) = 0.43
+ --- dist(#22,#1) = 0.519
+ --- dist(#23,#1) = 0.49
+ --- dist(#24,#1) = 0.502
+ --- dist(#25,#1) = 0.436
+ --- dist(#26,#1) = 0.468
+ --- dist(#27,#1) = 0.477
+ --- dist(#28,#1) = 0.498
+ --- dist(#29,#1) = 0.486
+ --- dist(#30,#1) = 0.423
++ Start refinement #3 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.88
+ - Smoothing source; radius=1.88
+ !source mask fill: ubot=64 usiz=159.5
+ - retaining old weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 32677 total points stored in 67 'TOHD(17.1973)' bloks (0 duplicates)
*[#26935=-0.23117] *[#26944=-0.232084] *[#26959=-0.2323] *[#26962=-0.232318] *[#26964=-0.232417] *[#26965=-0.232543] *[#26966=-0.232579] *[#26967=-0.232897] *[#26968=-0.233239] *[#26970=-0.233374] *[#26971=-0.233673] *[#26972=-0.233862] *[#26973=-0.234107] *[#26976=-0.234183] *[#26979=-0.234351] *[#26982=-0.234738] *[#26989=-0.234754] *[#26991=-0.234778] *[#26993=-0.234835] *[#26994=-0.234875] *[#26995=-0.23489] *[#26998=-0.234922] *[#27003=-0.234952] *[#27008=-0.23499] *[#27022=-0.235079] *[#27024=-0.235114] *[#27025=-0.235125] *[#27026=-0.235141] *[#27029=-0.235189] *[#27030=-0.235209] *[#27033=-0.235232] *[#27034=-0.235249] *[#27037=-0.235325] *[#27038=-0.235358] *[#27039=-0.235442] *[#27044=-0.235475] *[#27045=-0.235495] *[#27046=-0.235521] *[#27047=-0.235525] *[#27049=-0.235599] *[#27050=-0.235614] *[#27052=-0.235647] *[#27054=-0.235672]
+ - param set #1 has cost=-0.235672 [o=-1 t=-1]
+ -- Parameters = 86.4793 -3.3380 0.0217 -0.7549 -2.1405 -0.4338 0.9891 1.0267 0.9753 -0.0112 0.0131 -0.0443
+ - param set #2 has cost=-0.234666 [o=0 t=0]
+ -- Parameters = 86.4372 -3.2322 -0.0269 -0.6424 -1.6709 -0.2772 0.9885 1.0248 0.9691 -0.0071 0.0076 -0.0358
+ - param set #3 has cost=-0.025822 [o=42 t=21]
+ -- Parameters = 124.8635 -21.9185 -26.3384 8.9122 -2.8957 11.2486 0.9066 0.9500 0.9146 -0.0262 -0.0428 -0.0111
+ - param set #4 has cost=-0.024015 [o=2 t=2]
+ -- Parameters = 96.9853 -22.6084 36.5331 -10.5646 -42.3502 -25.8919 0.9936 0.9938 0.9909 -0.0015 0.0015 0.0018
+ - param set #5 has cost=-0.020677 [o=19 t=1]
+ -- Parameters = 120.2836 31.7782 37.8573 31.4903 -11.7839 -1.0432 1.0280 0.9925 0.9857 -0.0018 0.0016 0.0008
+ - param set #6 has cost=-0.021109 [o=11 t=4]
+ -- Parameters = 122.3854 3.2395 37.8890 32.1032 -27.2846 -8.9726 0.9996 1.0025 0.9997 -0.0002 -0.0004 0.0002
+ - param set #7 has cost=-0.021742 [o=81 t=18]
+ -- Parameters = 100.8685 15.8940 -35.1960 -31.0087 7.4956 -4.8791 0.9890 1.0230 0.9984 -0.0014 0.0064 -0.0026
+ - param set #8 has cost=-0.019976 [o=21 t=5]
+ -- Parameters = 115.3360 31.3654 37.9305 28.3074 -10.2193 2.1137 0.9882 0.9720 1.0138 0.0030 0.0001 -0.0012
+ - param set #9 has cost=-0.019055 [o=6 t=7]
+ -- Parameters = 124.6007 -28.5691 20.2957 5.1816 -41.5171 -44.5870 1.0131 0.9665 0.9992 -0.0041 -0.0017 -0.0082
+ - param set #10 has cost=-0.019574 [o=20 t=11]
+ -- Parameters = 99.3866 -29.8615 26.9020 -5.3338 -28.4230 -34.3867 1.0113 0.9855 1.0194 -0.0030 -0.0004 0.0010
+ - param set #11 has cost=-0.019180 [o=59 t=15]
+ -- Parameters = 120.9740 -8.5507 38.2855 35.6054 -28.5926 -2.1032 0.9445 0.9658 0.9799 0.0065 -0.0097 0.0127
+ - param set #12 has cost=-0.018566 [o=61 t=20]
+ -- Parameters = 95.1214 -16.0331 34.2759 18.9140 -38.5791 -27.1663 0.9916 1.0140 0.9960 0.0012 0.0011 -0.0035
+ - param set #13 has cost=-0.018186 [o=13 t=14]
+ -- Parameters = 94.6948 -15.8655 35.0391 15.9754 -38.3484 -28.1206 0.9895 1.0072 0.9927 -0.0019 0.0009 -0.0027
+ - param set #14 has cost=-0.018618 [o=12 t=24]
+ -- Parameters = 90.7229 19.1250 31.4046 30.8607 -12.3845 -26.7735 0.9996 0.9886 0.9229 -0.0090 0.0068 -0.0166
+ - param set #15 has cost=-0.018967 [o=27 t=3]
+ -- Parameters = 124.5882 -14.9891 37.9295 33.0540 -31.2298 -4.2381 1.0065 1.0007 0.9931 -0.0009 0.0053 0.0016
+ - param set #16 has cost=-0.017468 [o=8 t=25]
+ -- Parameters = 92.1824 -0.8246 28.2401 -37.9242 16.3884 -33.9570 0.9830 0.9798 0.9728 -0.0014 -0.0014 -0.0006
+ - param set #17 has cost=-0.018075 [o=70 t=17]
+ -- Parameters = 86.9585 -14.1999 -34.6419 -5.8658 10.1152 3.4199 0.9923 0.9807 0.9588 0.0087 0.0037 -0.0025
+ - param set #18 has cost=-0.016481 [o=87 t=8]
+ -- Parameters = 80.8196 10.8792 31.9415 39.1021 -23.3509 -12.5253 0.9963 0.9912 0.9875 -0.0009 -0.0018 0.0008
+ - param set #19 has cost=-0.017711 [o=3 t=27]
+ -- Parameters = 92.2159 -30.5634 24.8590 5.7157 -33.5922 -32.8620 0.9876 0.9841 0.9895 0.0194 0.0052 -0.0038
+ - param set #20 has cost=-0.021740 [o=76 t=6]
+ -- Parameters = 83.8861 4.6151 -39.5617 21.7046 10.7784 4.3335 1.0048 1.0034 1.0216 -0.0014 -0.0017 0.0062
+ - param set #21 has cost=-0.015678 [o=23 t=9]
+ -- Parameters = 88.9620 -11.8477 -34.5045 -12.1120 11.6847 4.7198 0.9995 0.9920 0.9895 0.0145 0.0006 -0.0002
+ - param set #22 has cost=-0.016589 [o=68 t=10]
+ -- Parameters = 94.9995 11.4811 -41.7072 -27.1686 18.0982 -14.7609 0.9953 1.0045 1.0033 0.0090 -0.0015 -0.0005
+ - param set #23 has cost=-0.016828 [o=33 t=16]
+ -- Parameters = 110.5268 17.5289 -34.6249 -44.8163 11.5456 22.9792 0.9797 1.0014 0.9661 -0.0008 0.0213 0.0032
+ - param set #24 has cost=-0.016103 [o=55 t=13]
+ -- Parameters = 101.9900 23.5558 -40.5738 -5.7638 -15.8114 40.9105 1.0031 0.9992 1.0094 0.0007 -0.0004 -0.0003
+ - param set #25 has cost=-0.014262 [o=26 t=12]
+ -- Parameters = 112.7557 -14.0757 -34.9144 -28.0999 24.1661 4.0230 0.9951 1.0018 1.0066 0.0048 0.0082 -0.0003
+ - param set #26 has cost=-0.012861 [o=24 t=23]
+ -- Parameters = 119.4894 -7.0076 -37.7482 -11.0534 5.0314 -12.8294 0.9963 1.0036 0.9832 -0.0072 0.0027 -0.0031
+ - param set #27 has cost=-0.017379 [o=43 t=26]
+ -- Parameters = 66.9122 8.7775 -38.5261 20.0114 18.3468 -6.1556 0.9969 0.9943 0.9711 0.0009 0.0066 0.0213
+ - param set #28 has cost=-0.012238 [o=15 t=22]
+ -- Parameters = 119.3286 0.1633 -40.0271 -13.9141 11.3075 -14.2199 0.9960 0.9968 1.0031 0.0023 -0.0007 -0.0006
+ - param set #29 has cost=-0.013300 [o=53 t=28]
+ -- Parameters = 118.9961 -8.2432 -38.8829 -34.5285 24.2673 -0.6308 1.0005 1.0040 1.0051 -0.0013 0.0007 0.0002
+ - param set #30 has cost=-0.012264 [o=74 t=19]
+ -- Parameters = 66.4348 14.9373 -34.0290 33.9352 20.6722 -10.4953 0.9986 1.0022 0.9993 0.0001 0.0001 0.0009
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0383
+ --- dist(#3,#1) = 0.48
+ --- dist(#4,#1) = 0.456
+ --- dist(#5,#1) = 0.44
+ --- dist(#6,#1) = 0.495
+ --- dist(#7,#1) = 0.473
+ --- dist(#8,#1) = 0.473
+ --- dist(#9,#1) = 0.474
+ --- dist(#10,#1) = 0.377
+ --- dist(#11,#1) = 0.478
+ --- dist(#12,#1) = 0.491
+ --- dist(#13,#1) = 0.476
+ --- dist(#14,#1) = 0.392
+ --- dist(#15,#1) = 0.428
+ --- dist(#16,#1) = 0.438
+ --- dist(#17,#1) = 0.433
+ --- dist(#18,#1) = 0.36
+ --- dist(#19,#1) = 0.413
+ --- dist(#20,#1) = 0.482
+ --- dist(#21,#1) = 0.49
+ --- dist(#22,#1) = 0.522
+ --- dist(#23,#1) = 0.443
+ --- dist(#24,#1) = 0.507
+ --- dist(#25,#1) = 0.432
+ --- dist(#26,#1) = 0.437
+ --- dist(#27,#1) = 0.486
+ --- dist(#28,#1) = 0.472
+ --- dist(#29,#1) = 0.426
+ --- dist(#30,#1) = 0.501
+ - Total coarse refinement net CPU time = 0.0 s; 9153 funcs
++ *** Fine pass begins ***
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.00
+ - Smoothing source; radius=1.00
+ !source mask fill: ubot=64 usiz=159.5
+ - retaining old weight image
+ * Exit alignment setup routine
++ Picking best parameter set out of 31 cases
+ 32726 total points stored in 68 'TOHD(17.1233)' bloks (0 duplicates)
+ - cost(#1)=-0.229302 * [o=-1 t=-1]
+ -- Parameters = 86.4793 -3.3380 0.0217 -0.7549 -2.1405 -0.4338 0.9891 1.0267 0.9753 -0.0112 0.0131 -0.0443
+ - cost(#2)=-0.228654 [o=0 t=0]
+ -- Parameters = 86.4372 -3.2322 -0.0269 -0.6424 -1.6709 -0.2772 0.9885 1.0248 0.9691 -0.0071 0.0076 -0.0358
+ - cost(#3)=-0.020796 [o=42 t=21]
+ -- Parameters = 124.8635 -21.9185 -26.3384 8.9122 -2.8957 11.2486 0.9066 0.9500 0.9146 -0.0262 -0.0428 -0.0111
+ - cost(#4)=-0.021017 [o=2 t=2]
+ -- Parameters = 96.9853 -22.6084 36.5331 -10.5646 -42.3502 -25.8919 0.9936 0.9938 0.9909 -0.0015 0.0015 0.0018
+ - cost(#5)=-0.019426 [o=81 t=18]
+ -- Parameters = 100.8685 15.8940 -35.1960 -31.0087 7.4956 -4.8791 0.9890 1.0230 0.9984 -0.0014 0.0064 -0.0026
+ - cost(#6)=-0.019844 [o=76 t=6]
+ -- Parameters = 83.8861 4.6151 -39.5617 21.7046 10.7784 4.3335 1.0048 1.0034 1.0216 -0.0014 -0.0017 0.0062
+ - cost(#7)=-0.018617 [o=11 t=4]
+ -- Parameters = 122.3854 3.2395 37.8890 32.1032 -27.2846 -8.9726 0.9996 1.0025 0.9997 -0.0002 -0.0004 0.0002
+ - cost(#8)=-0.016587 [o=19 t=1]
+ -- Parameters = 120.2836 31.7782 37.8573 31.4903 -11.7839 -1.0432 1.0280 0.9925 0.9857 -0.0018 0.0016 0.0008
+ - cost(#9)=-0.017151 [o=21 t=5]
+ -- Parameters = 115.3360 31.3654 37.9305 28.3074 -10.2193 2.1137 0.9882 0.9720 1.0138 0.0030 0.0001 -0.0012
+ - cost(#10)=-0.018519 [o=20 t=11]
+ -- Parameters = 99.3866 -29.8615 26.9020 -5.3338 -28.4230 -34.3867 1.0113 0.9855 1.0194 -0.0030 -0.0004 0.0010
+ - cost(#11)=-0.016436 [o=59 t=15]
+ -- Parameters = 120.9740 -8.5507 38.2855 35.6054 -28.5926 -2.1032 0.9445 0.9658 0.9799 0.0065 -0.0097 0.0127
+ - cost(#12)=-0.016901 [o=6 t=7]
+ -- Parameters = 124.6007 -28.5691 20.2957 5.1816 -41.5171 -44.5870 1.0131 0.9665 0.9992 -0.0041 -0.0017 -0.0082
+ - cost(#13)=-0.016877 [o=27 t=3]
+ -- Parameters = 124.5882 -14.9891 37.9295 33.0540 -31.2298 -4.2381 1.0065 1.0007 0.9931 -0.0009 0.0053 0.0016
+ - cost(#14)=-0.016763 [o=12 t=24]
+ -- Parameters = 90.7229 19.1250 31.4046 30.8607 -12.3845 -26.7735 0.9996 0.9886 0.9229 -0.0090 0.0068 -0.0166
+ - cost(#15)=-0.016364 [o=61 t=20]
+ -- Parameters = 95.1214 -16.0331 34.2759 18.9140 -38.5791 -27.1663 0.9916 1.0140 0.9960 0.0012 0.0011 -0.0035
+ - cost(#16)=-0.015450 [o=13 t=14]
+ -- Parameters = 94.6948 -15.8655 35.0391 15.9754 -38.3484 -28.1206 0.9895 1.0072 0.9927 -0.0019 0.0009 -0.0027
+ - cost(#17)=-0.016063 [o=70 t=17]
+ -- Parameters = 86.9585 -14.1999 -34.6419 -5.8658 10.1152 3.4199 0.9923 0.9807 0.9588 0.0087 0.0037 -0.0025
+ - cost(#18)=-0.015527 [o=3 t=27]
+ -- Parameters = 92.2159 -30.5634 24.8590 5.7157 -33.5922 -32.8620 0.9876 0.9841 0.9895 0.0194 0.0052 -0.0038
+ - cost(#19)=-0.015525 [o=8 t=25]
+ -- Parameters = 92.1824 -0.8246 28.2401 -37.9242 16.3884 -33.9570 0.9830 0.9798 0.9728 -0.0014 -0.0014 -0.0006
+ - cost(#20)=-0.016496 [o=43 t=26]
+ -- Parameters = 66.9122 8.7775 -38.5261 20.0114 18.3468 -6.1556 0.9969 0.9943 0.9711 0.0009 0.0066 0.0213
+ - cost(#21)=-0.014669 [o=33 t=16]
+ -- Parameters = 110.5268 17.5289 -34.6249 -44.8163 11.5456 22.9792 0.9797 1.0014 0.9661 -0.0008 0.0213 0.0032
+ - cost(#22)=-0.015020 [o=68 t=10]
+ -- Parameters = 94.9995 11.4811 -41.7072 -27.1686 18.0982 -14.7609 0.9953 1.0045 1.0033 0.0090 -0.0015 -0.0005
+ - cost(#23)=-0.014942 [o=87 t=8]
+ -- Parameters = 80.8196 10.8792 31.9415 39.1021 -23.3509 -12.5253 0.9963 0.9912 0.9875 -0.0009 -0.0018 0.0008
+ - cost(#24)=-0.014574 [o=55 t=13]
+ -- Parameters = 101.9900 23.5558 -40.5738 -5.7638 -15.8114 40.9105 1.0031 0.9992 1.0094 0.0007 -0.0004 -0.0003
+ - cost(#25)=-0.014193 [o=23 t=9]
+ -- Parameters = 88.9620 -11.8477 -34.5045 -12.1120 11.6847 4.7198 0.9995 0.9920 0.9895 0.0145 0.0006 -0.0002
+ - cost(#26)=-0.012148 [o=26 t=12]
+ -- Parameters = 112.7557 -14.0757 -34.9144 -28.0999 24.1661 4.0230 0.9951 1.0018 1.0066 0.0048 0.0082 -0.0003
+ - cost(#27)=-0.012000 [o=53 t=28]
+ -- Parameters = 118.9961 -8.2432 -38.8829 -34.5285 24.2673 -0.6308 1.0005 1.0040 1.0051 -0.0013 0.0007 0.0002
+ - cost(#28)=-0.011452 [o=24 t=23]
+ -- Parameters = 119.4894 -7.0076 -37.7482 -11.0534 5.0314 -12.8294 0.9963 1.0036 0.9832 -0.0072 0.0027 -0.0031
+ - cost(#29)=-0.011364 [o=74 t=19]
+ -- Parameters = 66.4348 14.9373 -34.0290 33.9352 20.6722 -10.4953 0.9986 1.0022 0.9993 0.0001 0.0001 0.0009
+ - cost(#30)=-0.010783 [o=15 t=22]
+ -- Parameters = 119.3286 0.1633 -40.0271 -13.9141 11.3075 -14.2199 0.9960 0.9968 1.0031 0.0023 -0.0007 -0.0006
+ - cost(#31)=-0.129342 [o=-2 t=-2]
+ -- Parameters = 86.1901 -3.4407 -1.7088 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
+ -num_rtb 99 ==> refine all 31 cases
+ - cost(#1)=-0.229705 * [o=-1 t=-1]
+ -- Parameters = 86.4786 -3.3794 0.0456 -0.7344 -2.0985 -0.4671 0.9912 1.0266 0.9763 -0.0110 0.0121 -0.0449
+ - cost(#2)=-0.229554 [o=0 t=0]
+ -- Parameters = 86.4108 -3.3549 0.0075 -0.6847 -1.9108 -0.2988 0.9897 1.0254 0.9729 -0.0093 0.0067 -0.0411
+ - cost(#3)=-0.020800 [o=42 t=21]
+ -- Parameters = 124.8687 -21.9275 -26.3385 8.9060 -2.8977 11.2572 0.9066 0.9499 0.9145 -0.0261 -0.0428 -0.0111
+ - cost(#4)=-0.021932 [o=2 t=2]
+ -- Parameters = 96.7132 -21.8547 37.0594 -11.0616 -41.9192 -26.4843 0.9931 0.9931 0.9950 -0.0013 0.0013 0.0069
+ - cost(#5)=-0.019536 [o=81 t=18]
+ -- Parameters = 100.8433 15.8919 -35.1646 -30.9880 7.4855 -4.8544 0.9870 1.0216 0.9993 -0.0015 0.0070 -0.0025
+ - cost(#6)=-0.021827 [o=76 t=6]
+ -- Parameters = 82.3364 4.8772 -39.6003 20.1755 10.4514 3.8632 1.0049 1.0059 1.0260 -0.0085 -0.0025 0.0079
+ - cost(#7)=-0.019391 [o=11 t=4]
+ -- Parameters = 122.1885 3.3850 37.7804 31.9993 -26.7764 -9.1048 1.0047 1.0043 1.0237 -0.0004 -0.0009 0.0010
+ - cost(#8)=-0.016887 [o=19 t=1]
+ -- Parameters = 120.3862 31.8744 37.9044 31.2181 -11.7938 -1.2006 1.0284 0.9884 0.9874 -0.0101 0.0025 0.0006
+ - cost(#9)=-0.017390 [o=21 t=5]
+ -- Parameters = 115.3469 31.2859 37.8570 28.2962 -10.3465 2.2030 0.9873 0.9648 1.0135 0.0016 0.0010 -0.0018
+ - cost(#10)=-0.018693 [o=20 t=11]
+ -- Parameters = 99.3262 -29.9447 26.9371 -5.3159 -28.4886 -34.0726 1.0107 0.9849 1.0213 -0.0036 -0.0003 0.0007
+ - cost(#11)=-0.016689 [o=59 t=15]
+ -- Parameters = 120.8994 -8.5654 38.2884 35.5660 -28.3844 -2.2898 0.9449 0.9652 0.9808 0.0065 -0.0097 0.0202
+ - cost(#12)=-0.017087 [o=6 t=7]
+ -- Parameters = 124.5712 -28.6409 20.3221 5.1349 -41.6487 -44.5124 1.0125 0.9652 0.9978 -0.0043 -0.0015 -0.0151
+ - cost(#13)=-0.018747 [o=27 t=3]
+ -- Parameters = 124.8220 -15.4132 35.7434 32.8513 -32.2128 -4.2913 1.0051 1.0020 0.9925 -0.0014 0.0061 0.0027
+ - cost(#14)=-0.016958 [o=12 t=24]
+ -- Parameters = 90.6424 19.0892 31.4461 30.8611 -12.4391 -26.9602 1.0001 0.9900 0.9213 -0.0096 0.0069 -0.0164
+ - cost(#15)=-0.016621 [o=61 t=20]
+ -- Parameters = 95.2265 -16.0288 34.3051 18.8769 -38.6949 -27.3742 0.9949 1.0116 0.9938 0.0095 0.0005 -0.0030
+ - cost(#16)=-0.015480 [o=13 t=14]
+ -- Parameters = 94.6942 -15.8722 35.0063 16.0018 -38.3548 -28.1013 0.9892 1.0073 0.9938 -0.0018 0.0006 -0.0023
+ - cost(#17)=-0.017580 [o=70 t=17]
+ -- Parameters = 87.0306 -14.2621 -34.6583 -9.0179 9.8610 3.8227 0.9879 0.9809 0.9564 0.0087 0.0030 -0.0024
+ - cost(#18)=-0.016272 [o=3 t=27]
+ -- Parameters = 92.1936 -30.6225 24.8890 5.7653 -33.6379 -32.8354 0.9906 0.9904 0.9919 0.0296 0.0046 -0.0038
+ - cost(#19)=-0.018831 [o=8 t=25]
+ -- Parameters = 92.0665 -2.7610 29.1632 -37.9380 15.4778 -34.5080 0.9731 0.9648 0.9439 -0.0023 -0.0035 0.0016
+ - cost(#20)=-0.016643 [o=43 t=26]
+ -- Parameters = 66.9145 8.7317 -38.5499 19.9377 18.3452 -6.1108 1.0030 0.9976 0.9713 0.0006 0.0051 0.0221
+ - cost(#21)=-0.015076 [o=33 t=16]
+ -- Parameters = 110.5064 17.5324 -34.6949 -44.8090 11.7870 22.7514 0.9787 1.0007 0.9652 -0.0008 0.0293 0.0042
+ - cost(#22)=-0.015064 [o=68 t=10]
+ -- Parameters = 94.9821 11.4850 -41.7087 -27.1684 18.0621 -14.7438 0.9953 1.0047 1.0033 0.0097 -0.0014 -0.0005
+ - cost(#23)=-0.015215 [o=87 t=8]
+ -- Parameters = 80.8903 10.8767 31.8345 38.9750 -23.2805 -12.6024 0.9964 0.9868 0.9912 -0.0075 -0.0002 0.0015
+ - cost(#24)=-0.014807 [o=55 t=13]
+ -- Parameters = 101.9003 23.5725 -40.5280 -5.6755 -15.6652 40.9313 1.0032 0.9991 1.0112 0.0085 -0.0004 -0.0001
+ - cost(#25)=-0.014391 [o=23 t=9]
+ -- Parameters = 88.9471 -11.7913 -34.3018 -12.1401 11.8714 4.7590 1.0001 0.9921 0.9903 0.0145 0.0001 -0.0000
+ - cost(#26)=-0.012283 [o=26 t=12]
+ -- Parameters = 112.7510 -14.0798 -34.9248 -28.0911 24.1450 4.0337 0.9952 1.0025 1.0059 0.0123 0.0083 -0.0004
+ - cost(#27)=-0.012050 [o=53 t=28]
+ -- Parameters = 119.0245 -8.2729 -38.8807 -34.4827 24.2436 -0.6653 1.0003 1.0042 1.0074 -0.0013 0.0015 0.0001
+ - cost(#28)=-0.011891 [o=24 t=23]
+ -- Parameters = 119.1164 -6.9467 -37.4876 -11.0812 5.1131 -12.3826 0.9973 1.0034 0.9823 -0.0065 0.0021 -0.0026
+ - cost(#29)=-0.011366 [o=74 t=19]
+ -- Parameters = 66.4278 14.9361 -34.0231 33.9244 20.6755 -10.4931 0.9986 1.0021 0.9994 0.0001 0.0002 0.0008
+ - cost(#30)=-0.012654 [o=15 t=22]
+ -- Parameters = 118.6806 -0.2779 -39.9209 -10.4236 10.9901 -14.6070 0.9859 0.9982 1.0056 0.0036 -0.0026 0.0003
+ - cost(#31)=-0.226142 [o=-2 t=-2]
+ -- Parameters = 86.2853 -3.2417 0.0041 -0.6043 -1.3783 0.2327 0.9846 1.0247 0.9738 -0.0073 -0.0072 -0.0283
+ - case #1 [o=-1 t=-1] is now the best
+ - Initial cost = -0.229705
+ - Initial fine Parameters = 86.4786 -3.3794 0.0456 -0.7344 -2.0985 -0.4671 0.9912 1.0266 0.9763 -0.0110 0.0121 -0.0449
+ - Finalish cost = -0.230087 ; 443 funcs
+ - ini Finalish Parameters = 86.4243 -3.3934 0.0579 -0.6907 -2.4008 -0.2686 0.9907 1.0261 0.9755 -0.0090 0.0076 -0.0512
+ - Final cost = -0.230087 ; 283 funcs
+ Final fine fit Parameters:
x-shift= 86.4239 y-shift= -3.3912 z-shift= 0.0590 ... enorm= 86.4904 mm
z-angle= -0.6917 x-angle= -2.3988 y-angle= -0.2681 ... total= 2.5094 deg
x-scale= 0.9907 y-scale= 1.0261 z-scale= 0.9755 ... vol3D= 0.9917 = base bigger than source
y/x-shear= -0.0090 z/x-shear= 0.0076 z/y-shear= -0.0512
+ - Fine net CPU time = 0.0 s
++ Computing output image
++ image warp: parameters = 86.4239 -3.3912 0.0590 -0.6917 -2.3988 -0.2681 0.9907 1.0261 0.9755 -0.0090 0.0076 -0.0512
++ Unloading unneeded data
++ Output dataset ./sub-08_T1w_al_junk+orig.BRIK
++ Wrote -1Dmatrix_save ./sub-08_T1w_al_junk_mat.aff12.1D
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 17.2
++ ###########################################################
++ # PLEASE check results VISUALLY for alignment quality #
++ ###########################################################
#++ Creating final output: skullstripped anat data
copying from dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_processing/sub_08.results/__tt_sub-08_T1w_ns+orig to /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_processing/sub_08.results/sub-08_T1w_ns+orig
#Script is running (command trimmed):
3dcopy ./__tt_sub-08_T1w_ns+orig sub-08_T1w_ns
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ Creating final output: anat data aligned to epi
# copy is not necessary
#++ Saving history
#Script is running (command trimmed):
3dNotes -h "align_epi_anat.py -anat2epi -anat sub-08_T1w+orig \
-save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base \
0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off" \
./sub-08_T1w_al_junk+orig
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-08_T1w*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
# Finished alignment successfully
@auto_tlrc -base MNI_avg152T1+tlrc -input sub-08_T1w_ns+orig -no_ss
*********** Warning *************
Dataset centers are 92.736319 mm
apart. If registration fails, or if
parts of the original anatomy gets
cropped, try adding option
-init_xform AUTO_CENTER
to your @auto_tlrc command.
*********************************
Padding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp__ref_MNI_avg152T1_15pad+tlrc.BRIK
Resampling ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp_step+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp+tlrc.BRIK
Clipping -0.000100 1336.000100 ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./__ats_tmp___rs_sub-08_T1w_ns+tlrc.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___rs_sub-08_T1w_ns+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_NN+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_edge_art+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Registration (cubic final interpolation) ...
++ 3dWarpDrive: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Initial scale factor set to 0.30/129.00=4.3e+02
RMS[0] = 0.505204 0.228999 ITER = 15/50
0.505204
Warping has converged.
mv: ‘sub-08_T1w_ns.Xaff12.1D’ and ‘sub-08_T1w_ns.Xaff12.1D’ are the same file
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Applying brain mask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__reg_braintlrcstep+orig.BRIK
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename __ats_tmp___pad15_sub-08_T1w_ns+orig.HEAD -> __ats_tmp___pad15_sub-08_T1w_ns.skl+orig.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___pad15_sub-08_T1w_ns+orig.BRIK -> __ats_tmp___pad15_sub-08_T1w_ns.skl+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp___pad15_sub-08_T1w_ns+orig.BRIK
Unpadding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp___upad15_sub-08_T1w_ns+orig.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_sub-08_T1w_ns+orig
*+ WARNING: Changing the space of an ORIG view dataset may cause confusion!
*+ WARNING: NIFTI copies will be interpreted as TLRC view (not TLRC space).
*+ WARNING: Consider changing the view of the dataset to TLRC view also
++ 3drefit processed 1 datasets
Changing view of transformed anatomy
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_sub-08_T1w_ns+orig.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Setting parent with 3drefit -wset sub-08_T1w_ns+orig __ats_tmp___upad15_sub-08_T1w_ns+tlrc
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_sub-08_T1w_ns+tlrc
+ setting Warp parent
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Warning: ignoring +tlrc on new_prefix.
++ THD_rename_dataset_files: rename __ats_tmp___upad15_sub-08_T1w_ns+tlrc.HEAD -> sub-08_T1w_ns+tlrc.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___upad15_sub-08_T1w_ns+tlrc.BRIK -> sub-08_T1w_ns+tlrc.BRIK
Cleanup ...
cat_matvec sub-08_T1w_ns+tlrc::WARP_DATA -I
if ( ! -f sub-08_T1w_ns+tlrc.HEAD ) then
foreach run ( 01 02 )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r01.1D -prefix rm.epi.volreg.r01 -cubic -1Dmatrix_save mat.r01.vr.aff12.1D pb01.sub_08.r01.tshift+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb01.sub_08.r01.tshift+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ Automask has 40918 voxels
+ 5959 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.014 pitch=-0.044 yaw=-0.059 dS=-0.092 dL=-0.041 dP=-0.083
++ Mean: roll=+0.022 pitch=+0.035 yaw=+0.065 dS=+0.016 dL=+0.029 dP=+0.013
++ Max : roll=+0.119 pitch=+0.184 yaw=+0.253 dS=+0.181 dL=+0.123 dP=+0.235
++ Max displacements (mm) for each sub-brick:
0.15(0.00) 0.09(0.16) 0.06(0.09) 0.05(0.08) 0.07(0.08) 0.07(0.03) 0.08(0.05) 0.06(0.05) 0.15(0.13) 0.09(0.12) 0.15(0.12) 0.09(0.13) 0.11(0.10) 0.09(0.06) 0.09(0.05) 0.11(0.04) 0.04(0.10) 0.08(0.08) 0.07(0.05) 0.13(0.10) 0.06(0.11) 0.06(0.09) 0.07(0.09) 0.03(0.05) 0.09(0.07) 0.06(0.13) 0.07(0.09) 0.05(0.08) 0.00(0.05) 0.06(0.06) 0.06(0.08) 0.05(0.06) 0.10(0.07) 0.06(0.06) 0.07(0.09) 0.06(0.10) 0.08(0.08) 0.08(0.07) 0.12(0.10) 0.08(0.11) 0.09(0.08) 0.11(0.07) 0.05(0.08) 0.08(0.05) 0.03(0.08) 0.07(0.06) 0.08(0.09) 0.06(0.05) 0.10(0.07) 0.08(0.09) 0.12(0.10) 0.10(0.08) 0.06(0.07) 0.14(0.14) 0.09(0.11) 0.11(0.10) 0.14(0.06) 0.09(0.12) 0.12(0.11) 0.10(0.09) 0.10(0.06) 0.15(0.07) 0.09(0.12) 0.11(0.10) 0.09(0.15) 0.14(0.16) 0.09(0.09) 0.10(0.03) 0.11(0.07) 0.08(0.07) 0.13(0.11) 0.09(0.11) 0.13(0.11) 0.09(0.08) 0.10(0.06) 0.13(0.10) 0.10(0.09) 0.14(0.11) 0.11(0.14) 0.11(0.10) 0.09(0.07) 0.10(0.08) 0.11(0.06) 0.10(0.10) 0.16(0.16) 0.10(0.16) 0.13(0.14) 0.13(0.11) 0.17(0.07) 0.18(0.07) 0.15(0.09) 0.15(0.07) 0.12(0.10) 0.17(0.10) 0.15(0.05) 0.16(0.07) 0.14(0.06) 0.16(0.10) 0.24(0.15) 0.14(0.17) 0.21(0.14) 0.16(0.12) 0.29(0.22) 0.20(0.16) 0.23(0.11) 0.21(0.07) 0.21(0.10) 0.23(0.05) 0.19(0.12) 0.22(0.08) 0.18(0.09) 0.23(0.10) 0.19(0.11) 0.28(0.14) 0.21(0.10) 0.23(0.06) 0.23(0.07) 0.29(0.15) 0.22(0.17) 0.29(0.10) 0.30(0.04) 0.27(0.07) 0.31(0.08) 0.28(0.06) 0.34(0.11) 0.30(0.07) 0.37(0.11) 0.28(0.15) 0.36(0.14) 0.29(0.14) 0.36(0.15) 0.32(0.30) 0.41(0.27) 0.49(0.29) 0.46(0.11) 0.49(0.18) 0.56(0.18) 0.55(0.14) 0.55(0.10) 0.54(0.03) 0.52(0.12) 0.51(0.14) 0.48(0.15) 0.41(0.17) 0.41(0.22) 0.42(0.12)
++ Max displacement in automask = 0.56 (mm) at sub-brick 136
++ Max delta displ in automask = 0.30 (mm) at sub-brick 131
++ Wrote dataset to disk in ./rm.epi.volreg.r01+orig.BRIK
3dcalc -overwrite -a pb01.sub_08.r01.tshift+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-08_T1w_ns+tlrc::WARP_DATA -I sub-08_T1w_al_junk_mat.aff12.1D -I mat.r01.vr.aff12.1D
3dAllineate -base sub-08_T1w_ns+tlrc -input pb01.sub_08.r01.tshift+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb01.sub_08.r01.tshift+orig.HEAD
++ Base dataset: ./sub-08_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 1.177 -7.574 -29.220
+ shift search range is +/- = 57.780 69.336 57.780
+ 2.0% 10.9% 50.6%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.6
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-08_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 2.430 -17.988 -31.461
+ shift search range is +/- = 57.780 69.336 57.780
+ 4.2% 25.9% 54.4%
++ Output dataset ./rm.epi.1.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.1
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r01 rm.epi.1.r01+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.epi.min.r01+tlrc.BRIK
end
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r02.1D -prefix rm.epi.volreg.r02 -cubic -1Dmatrix_save mat.r02.vr.aff12.1D pb01.sub_08.r02.tshift+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb01.sub_08.r02.tshift+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ Automask has 40888 voxels
+ 5952 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=+0.065 pitch=-0.196 yaw=-0.295 dS=-0.284 dL=+0.045 dP=-0.220
++ Mean: roll=+0.160 pitch=+0.114 yaw=+0.164 dS=-0.032 dL=+0.132 dP=-0.070
++ Max : roll=+0.233 pitch=+0.250 yaw=+0.335 dS=+0.142 dL=+0.306 dP=+0.067
++ Max displacements (mm) for each sub-brick:
0.70(0.00) 0.61(0.16) 0.62(0.09) 0.60(0.07) 0.58(0.06) 0.63(0.08) 0.61(0.07) 0.62(0.08) 0.60(0.07) 0.55(0.07) 0.62(0.08) 0.61(0.11) 0.61(0.07) 0.59(0.05) 0.50(0.10) 0.54(0.06) 0.52(0.08) 0.57(0.09) 0.58(0.09) 0.56(0.09) 0.58(0.06) 0.56(0.07) 0.59(0.10) 0.57(0.09) 0.57(0.07) 0.59(0.05) 0.56(0.05) 0.59(0.08) 0.60(0.08) 0.58(0.07) 0.58(0.07) 0.58(0.06) 0.58(0.02) 0.59(0.05) 0.57(0.34) 0.38(0.39) 0.47(0.48) 0.90(0.47) 0.59(0.35) 0.49(0.19) 0.50(0.16) 0.46(0.12) 0.48(0.16) 0.46(0.17) 0.48(0.13) 0.47(0.10) 0.48(0.10) 0.49(0.09) 0.51(0.10) 0.53(0.06) 0.51(0.09) 0.53(0.09) 0.50(0.10) 0.50(0.09) 0.50(0.08) 0.49(0.07) 0.52(0.06) 0.46(0.10) 0.46(0.21) 0.52(0.33) 0.50(0.20) 0.54(0.12) 0.54(0.09) 0.60(0.12) 0.55(0.14) 0.59(0.12) 0.58(0.10) 0.62(0.13) 0.53(0.11) 0.49(0.14) 0.49(0.04) 0.46(0.07) 0.55(0.11) 0.48(0.14) 0.57(0.15) 0.46(0.14) 0.54(0.11) 0.44(0.13) 0.51(0.11) 0.47(0.10) 0.55(0.15) 0.59(0.11) 0.56(0.13) 0.53(0.13) 0.60(0.14) 0.58(0.10) 0.55(0.04) 0.53(0.07) 0.59(0.11) 0.49(0.15) 0.57(0.14) 0.52(0.17) 0.77(0.46) 0.58(0.27) 0.57(0.20) 0.57(0.10) 0.58(0.13) 0.56(0.14) 0.57(0.14) 0.55(0.09) 0.60(0.10) 0.55(0.09) 0.55(0.05) 0.55(0.04) 0.57(0.07) 0.61(0.06) 0.53(0.11) 0.67(0.16) 0.55(0.16) 0.59(0.11) 0.53(0.10) 0.54(0.06) 0.57(0.07) 0.51(0.12) 0.56(0.12) 0.57(0.17) 0.64(0.20) 0.58(0.21) 0.56(0.09) 0.60(0.07) 0.55(0.07) 0.61(0.08) 0.56(0.12) 0.60(0.14) 0.59(0.10) 0.61(0.07) 0.59(0.06) 0.56(0.08) 0.67(0.14) 0.61(0.12) 0.64(0.14) 0.64(0.04) 0.64(0.19) 0.57(0.25) 0.54(0.10) 0.54(0.07) 0.53(0.05) 0.57(0.11) 0.53(0.11) 0.58(0.12) 0.53(0.09) 0.55(0.06) 0.52(0.11) 0.56(0.13) 0.55(0.10) 0.53(0.06)
++ Max displacement in automask = 0.90 (mm) at sub-brick 37
++ Max delta displ in automask = 0.48 (mm) at sub-brick 36
++ Wrote dataset to disk in ./rm.epi.volreg.r02+orig.BRIK
3dcalc -overwrite -a pb01.sub_08.r02.tshift+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-08_T1w_ns+tlrc::WARP_DATA -I sub-08_T1w_al_junk_mat.aff12.1D -I mat.r02.vr.aff12.1D
3dAllineate -base sub-08_T1w_ns+tlrc -input pb01.sub_08.r02.tshift+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb01.sub_08.r02.tshift+orig.HEAD
++ Base dataset: ./sub-08_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 1.104 -7.508 -29.208
+ shift search range is +/- = 57.780 69.336 57.780
+ 1.9% 10.8% 50.6%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.6
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-08_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 2.430 -17.988 -31.461
+ shift search range is +/- = 57.780 69.336 57.780
+ 4.2% 25.9% 54.4%
++ Output dataset ./rm.epi.1.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.1
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r02 rm.epi.1.r02+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.epi.min.r02+tlrc.BRIK
end
cat dfile.r01.1D dfile.r02.1D
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -derivative -collapse_cols euclidean_norm -write motion_sub_08_enorm.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dMean -datum short -prefix rm.epi.mean rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.HEAD
++ 3dMean: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dcalc -a rm.epi.mean+tlrc -expr step(a-0.999) -prefix mask_epi_extents
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./mask_epi_extents+tlrc.BRIK
foreach run ( 01 02 )
3dcalc -a rm.epi.nomask.r01+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb02.sub_08.r01.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb02.sub_08.r01.volreg+tlrc.BRIK
end
3dcalc -a rm.epi.nomask.r02+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb02.sub_08.r02.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb02.sub_08.r02.volreg+tlrc.BRIK
end
cat_matvec -ONELINE sub-08_T1w_ns+tlrc::WARP_DATA -I sub-08_T1w_al_junk_mat.aff12.1D -I
3dAllineate -base sub-08_T1w_ns+tlrc -input vr_base_min_outlier+orig -1Dmatrix_apply mat.basewarp.aff12.1D -mast_dxyz 3 -prefix final_epi_vr_base_min_outlier
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./vr_base_min_outlier+orig.HEAD
++ Base dataset: ./sub-08_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 1.223 -7.586 -29.179
+ shift search range is +/- = 57.780 69.336 57.780
+ 2.1% 10.9% 50.5%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./final_epi_vr_base_min_outlier+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dcopy sub-08_T1w_ns+tlrc anat_final.sub_08
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dAllineate -base final_epi_vr_base_min_outlier+tlrc -allcostX -input anat_final.sub_08+tlrc
tee out.allcostX.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./anat_final.sub_08+tlrc.HEAD
++ Base dataset: ./final_epi_vr_base_min_outlier+tlrc.HEAD
++ Loading datasets into memory
++ Local correlation: blok type = 'TOHD(15.531)'
+ -cmass x y z shifts = -13.464 0.425 2.519
+ shift search range is +/- = 75.114 82.818 67.410
*+ WARNING: No output dataset will be calculated
++ OpenMP thread count = 15
+ initial Parameters = 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ allcost output: init #0
ls = 0.230214
sp = 0.332702
mi = 3.12782
crM = 0.135544
nmi = 0.87499
je = 3.12782
hel = -0.085955
crA = 0.36856
crU = 0.385647
lss = 0.769786
lpc = 0.447039
lpa = 0.552961
lpc+ = 0.680087
lpa+ = 0.786009
++ Output dataset ./volumized+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.8
++ ###########################################################
3dAllineate -source sub-08_T1w+orig -master anat_final.sub_08+tlrc -final wsinc5 -1Dmatrix_apply warp.anat.Xat.1D -prefix anat_w_skull_warped
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ -1Dmatrix_apply: converting input 3x4 array to 1 row of 12 numbers
++ Source dataset: ./sub-08_T1w+orig.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 56.175 81.855 81.855
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset ./anat_w_skull_warped+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.4
++ ###########################################################
foreach run ( 01 02 )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb03.sub_08.r01.blur pb02.sub_08.r01.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb03.sub_08.r01.blur+tlrc.BRIK
end
3dmerge -1blur_fwhm 4.0 -doall -prefix pb03.sub_08.r02.blur pb02.sub_08.r02.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb03.sub_08.r02.blur+tlrc.BRIK
end
foreach run ( 01 02 )
3dAutomask -prefix rm.mask_r01 pb03.sub_08.r01.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb03.sub_08.r01.blur+tlrc
++ Forming automask
+ Fixed clip level = 343.984131
+ Used gradual clip level = 319.658783 .. 361.950439
+ Number voxels above clip level = 95016
+ Clustering voxels ...
+ Largest cluster has 94183 voxels
+ Clustering voxels ...
+ Largest cluster has 94053 voxels
+ Filled 136 voxels in small holes; now have 94189 voxels
+ Clustering voxels ...
+ Largest cluster has 94189 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 177444 voxels
+ Mask now has 94189 voxels
++ 94189 voxels in the mask [out of 271633: 34.68%]
++ first 0 x-planes are zero [from L]
++ last 2 x-planes are zero [from R]
++ first 5 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 9 z-planes are zero [from S]
++ Output dataset ./rm.mask_r01+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dAutomask -prefix rm.mask_r02 pb03.sub_08.r02.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb03.sub_08.r02.blur+tlrc
++ Forming automask
+ Fixed clip level = 342.683807
+ Used gradual clip level = 318.450317 .. 360.525879
+ Number voxels above clip level = 95030
+ Clustering voxels ...
+ Largest cluster has 94190 voxels
+ Clustering voxels ...
+ Largest cluster has 94071 voxels
+ Filled 130 voxels in small holes; now have 94201 voxels
+ Clustering voxels ...
+ Largest cluster has 94201 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 177432 voxels
+ Mask now has 94201 voxels
++ 94201 voxels in the mask [out of 271633: 34.68%]
++ first 0 x-planes are zero [from L]
++ last 2 x-planes are zero [from R]
++ first 5 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 9 z-planes are zero [from S]
++ Output dataset ./rm.mask_r02+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dmask_tool -inputs rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.HEAD -union -prefix full_mask.sub_08
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 0 over 2 volumes gives min count 0
++ voxel limits: 0 clipped, 94280 survived, 177353 were zero
++ writing result full_mask.sub_08...
++ Output dataset ./full_mask.sub_08+tlrc.BRIK
3dresample -master full_mask.sub_08+tlrc -input sub-08_T1w_ns+tlrc -prefix rm.resam.anat
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.anat+tlrc -prefix mask_anat.sub_08
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 81218 survived, 190415 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_anat.sub_08...
++ Output dataset ./mask_anat.sub_08+tlrc.BRIK
3dmask_tool -input full_mask.sub_08+tlrc mask_anat.sub_08+tlrc -inter -prefix mask_epi_anat.sub_08
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 1 over 2 volumes gives min count 2
++ voxel limits: 32128 clipped, 71685 survived, 167820 were zero
++ writing result mask_epi_anat.sub_08...
++ Output dataset ./mask_epi_anat.sub_08+tlrc.BRIK
3dABoverlap -no_automask full_mask.sub_08+tlrc mask_anat.sub_08+tlrc
tee out.mask_ae_overlap.txt
++ 3dABoverlap: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#A=./full_mask.sub_08+tlrc.BRIK B=./mask_anat.sub_08+tlrc.BRIK
#A #B #(A uni B) #(A int B) #(A \ B) #(B \ A) %(A \ B) %(B \ A) Rx(B/A) Ry(B/A) Rz(B/A)
94280 81218 103813 71685 22595 9533 23.9658 11.7375 0.6653 0.9754 1.0909
3ddot -dodice full_mask.sub_08+tlrc mask_anat.sub_08+tlrc
tee out.mask_ae_dice.txt
0.816932
3dresample -master full_mask.sub_08+tlrc -prefix ./rm.resam.group -input /opt/afni-latest/MNI_avg152T1+tlrc
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.group+tlrc -prefix mask_group
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_group...
++ Output dataset ./mask_group+tlrc.BRIK
3ddot -dodice mask_anat.sub_08+tlrc mask_group+tlrc
tee out.mask_at_dice.txt
0.888823
foreach run ( 01 02 )
3dTstat -prefix rm.mean_r01 pb03.sub_08.r01.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.mean_r01+tlrc.BRIK
3dcalc -a pb03.sub_08.r01.blur+tlrc -b rm.mean_r01+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb04.sub_08.r01.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb04.sub_08.r01.scale+tlrc.BRIK
end
3dTstat -prefix rm.mean_r02 pb03.sub_08.r02.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.mean_r02+tlrc.BRIK
3dcalc -a pb03.sub_08.r02.blur+tlrc -b rm.mean_r02+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb04.sub_08.r02.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb04.sub_08.r02.scale+tlrc.BRIK
end
gen_epi_review.py -script @epi_review.sub_08 -dsets pb00.sub_08.r01.tcat+orig.HEAD pb00.sub_08.r02.tcat+orig.HEAD
rm -f rm.epi.1.r01+tlrc.BRIK rm.epi.1.r01+tlrc.HEAD rm.epi.1.r02+tlrc.BRIK rm.epi.1.r02+tlrc.HEAD rm.epi.all1+orig.BRIK rm.epi.all1+orig.HEAD rm.epi.mean+tlrc.BRIK rm.epi.mean+tlrc.HEAD rm.epi.min.r01+tlrc.BRIK rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.BRIK rm.epi.min.r02+tlrc.HEAD rm.epi.nomask.r01+tlrc.BRIK rm.epi.nomask.r01+tlrc.HEAD rm.epi.nomask.r02+tlrc.BRIK rm.epi.nomask.r02+tlrc.HEAD rm.epi.volreg.r01+orig.BRIK rm.epi.volreg.r01+orig.HEAD rm.epi.volreg.r02+orig.BRIK rm.epi.volreg.r02+orig.HEAD rm.mask_r01+tlrc.BRIK rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.BRIK rm.mask_r02+tlrc.HEAD rm.mean_r01+tlrc.BRIK rm.mean_r01+tlrc.HEAD rm.mean_r02+tlrc.BRIK rm.mean_r02+tlrc.HEAD rm.resam.anat+tlrc.BRIK rm.resam.anat+tlrc.HEAD rm.resam.group+tlrc.BRIK rm.resam.group+tlrc.HEAD
if ( -e @ss_review_basic ) then
cd ..
echo execution finished: `date`
date
execution finished: Tue Nov 4 03:35:52 UTC 2025
🔔 While running, pay attention to the following warnings:
3dTshift Warning:
+ WARNING: dataset is already aligned in time!
+ WARNING: ==>> output dataset is just a copy of input dataset
✅ No action needed — this simply means no time-shifting was necessary. You could re-analyze this data by omitting both the 3dTcat and 3dTshift preprocessing steps, and it would get the same result.
@auto_tlrc Warning:
*********** Warning *************
Dataset centers are 92.736319 mm
apart. If registration fails, or if
parts of the original anatomy gets
cropped, try adding option
-init_xform AUTO_CENTER
to your @auto_tlrc command.
*********************************
⚠️ This warning requires fixing: we need to modify the proc.sub_08 script to add -init_xform AUTO_CENTER to the @auto_tlrc command to improve alignment.
# Edit proc.sub_08
with open('./proc.sub_08', 'r') as file:
lines = file.readlines()
# Insert "-init_xform AUTO_CENTER" into the @auto_tlrc command
for idx, line in enumerate(lines):
if line.strip().startswith('@auto_tlrc'):
lines[idx] = line.strip() + ' -init_xform AUTO_CENTER\n'
# Save it back
with open('proc.sub_08', 'w') as file:
file.writelines(lines)
# Delete any previous output (if needed)
! rm -r ./afni_processing/sub_08.results
# Rerun the processing script
! tcsh -xef proc.sub_08 |& tee output.proc.sub_08
echo auto-generated by afni_proc.py, Tue Nov 4 03:31:50 2025
auto-generated by afni_proc.py, Tue Nov 4 03:31:50 2025
echo (version 7.16, May 19, 2021)
(version 7.16, May 19, 2021)
echo execution started: `date`
date
execution started: Tue Nov 4 03:35:54 UTC 2025
afni -ver
Precompiled binary linux_openmp_64: Jul 8 2021 (Version AFNI_21.2.00 'Nerva')
afni_history -check_date 27 Jun 2019
-- is current: afni_history as new as: 27 Jun 2019
most recent entry is: 30 Jun 2021
if ( 0 ) then
if ( 0 > 0 ) then
set subj = sub_08
endif
set output_dir = ./afni_processing/sub_08.results
if ( -d ./afni_processing/sub_08.results ) then
set runs = ( `count -digits 2 1 2` )
count -digits 2 1 2
mkdir -p ./afni_processing/sub_08.results
mkdir ./afni_processing/sub_08.results/stimuli
3dcopy ds000102/sub-08/anat/sub-08_T1w.nii.gz ./afni_processing/sub_08.results/sub-08_T1w
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dTcat -prefix ./afni_processing/sub_08.results/pb00.sub_08.r01.tcat ds000102/sub-08/func/sub-08_task-flanker_run-1_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.7 s
3dTcat -prefix ./afni_processing/sub_08.results/pb00.sub_08.r02.tcat ds000102/sub-08/func/sub-08_task-flanker_run-2_bold.nii.gz[0..$]
++ 3dTcat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ elapsed time = 0.7 s
set tr_counts = ( 146 146 )
cd ./afni_processing/sub_08.results
touch out.pre_ss_warn.txt
foreach run ( 01 02 )
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub_08.r01.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 38915 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r01.1D{0} -expr step(a-0.4)
end
3dToutcount -automask -fraction -polort 2 -legendre pb00.sub_08.r02.tcat+orig
++ 3dToutcount: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 38896 voxels passed mask/clip
if ( `1deval -a outcount.r$run.1D"{0}" -expr "step(a-0.4)"` ) then
1deval -a outcount.r02.1D{0} -expr step(a-0.4)
end
cat outcount.r01.1D outcount.r02.1D
set minindex = `3dTstat -argmin -prefix - outcount_rall.1D\'`
3dTstat -argmin -prefix - outcount_rall.1D'
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
*+ WARNING: Input dataset is not 3D+time; assuming TR=1.0
set ovals = ( `1d_tool.py -set_run_lengths $tr_counts
-index_to_run_tr $minindex` )
1d_tool.py -set_run_lengths 146 146 -index_to_run_tr 28
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
set minoutrun = 01
set minouttr = 28
echo min outlier: run 01, TR 28
tee out.min_outlier.txt
min outlier: run 01, TR 28
foreach run ( 01 02 )
3dTshift -tzero 0 -quintic -prefix pb01.sub_08.r01.tshift pb00.sub_08.r01.tcat+orig
++ 3dTshift: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: dataset is already aligned in time!
*+ WARNING: ==>> output dataset is just a copy of input dataset
end
3dTshift -tzero 0 -quintic -prefix pb01.sub_08.r02.tshift pb00.sub_08.r02.tcat+orig
++ 3dTshift: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: dataset is already aligned in time!
*+ WARNING: ==>> output dataset is just a copy of input dataset
end
3dbucket -prefix vr_base_min_outlier pb01.sub_08.r01.tshift+orig[28]
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
align_epi_anat.py -anat2epi -anat sub-08_T1w+orig -save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base 0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off
#++ align_epi_anat version: 1.62
#++ turning off volume registration
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./vr_base_min_outlier+orig
#Script is running (command trimmed):
3dAttribute DELTA ./sub-08_T1w+orig
#++ Multi-cost is lpc
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-08_T1w*
#Script is running (command trimmed):
3dcopy ./sub-08_T1w+orig ./__tt_sub-08_T1w+orig
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ Removing skull from anat data
#Script is running (command trimmed):
3dSkullStrip -orig_vol -input ./__tt_sub-08_T1w+orig -prefix ./__tt_sub-08_T1w_ns
#Script is running (command trimmed):
3dinfo ./__tt_sub-08_T1w_ns+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_processing/sub_08.results/__tt_sub-08_T1w_ns+orig is not oblique
#Script is running (command trimmed):
3dinfo ./vr_base_min_outlier+orig | \grep 'Data Axes Tilt:'|\grep 'Oblique'
#++ Dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_processing/sub_08.results/vr_base_min_outlier+orig is not oblique
#++ using 0th sub-brick because only one found
#Script is running (command trimmed):
3dbucket -prefix ./__tt_vr_base_min_outlier_ts ./vr_base_min_outlier+orig'[0]'
++ 3dbucket: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ removing skull or area outside brain
#Script is running (command trimmed):
3dAutomask -apply_prefix ./__tt_vr_base_min_outlier_ts_ns ./__tt_vr_base_min_outlier_ts+orig
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset ./__tt_vr_base_min_outlier_ts+orig
++ Forming automask
+ Fixed clip level = 345.050415
+ Used gradual clip level = 330.013794 .. 365.519012
+ Number voxels above clip level = 39471
+ Clustering voxels ...
+ Largest cluster has 38949 voxels
+ Clustering voxels ...
+ Largest cluster has 38549 voxels
+ Filled 354 voxels in small holes; now have 38903 voxels
+ Filled 3 voxels in large holes; now have 38906 voxels
+ Clustering voxels ...
+ Largest cluster has 38906 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 124934 voxels
+ Mask now has 38906 voxels
++ 38906 voxels in the mask [out of 163840: 23.75%]
++ first 11 x-planes are zero [from L]
++ last 11 x-planes are zero [from R]
++ first 0 y-planes are zero [from P]
++ last 5 y-planes are zero [from A]
++ first 1 z-planes are zero [from I]
++ last 3 z-planes are zero [from S]
++ applying mask to original data
++ Writing masked data
++ Output dataset ./__tt_vr_base_min_outlier_ts_ns+orig.BRIK
++ CPU time = 0.000000 sec
#++ Computing weight mask
#Script is running (command trimmed):
3dBrickStat -automask -percentile 90.000000 1 90.000000 ./__tt_vr_base_min_outlier_ts_ns+orig
#++ Applying threshold of 819.000000 on /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_processing/sub_08.results/__tt_vr_base_min_outlier_ts_ns+orig
#Script is running (command trimmed):
3dcalc -datum float -prefix ./__tt_vr_base_min_outlier_ts_ns_wt -a ./__tt_vr_base_min_outlier_ts_ns+orig -expr 'min(1,(a/819.000000))'
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__tt_vr_base_min_outlier_ts_ns_wt+orig.BRIK
#++ Aligning anat data to epi data
#Script is running (command trimmed):
3dAllineate -lpc -wtprefix ./__tt_sub-08_T1w_ns_al_junk_wtal -weight ./__tt_vr_base_min_outlier_ts_ns_wt+orig -source ./__tt_sub-08_T1w_ns+orig -prefix ./sub-08_T1w_al_junk -base ./__tt_vr_base_min_outlier_ts_ns+orig -cmass -1Dmatrix_save ./sub-08_T1w_al_junk_mat.aff12.1D -master BASE -mast_dxyz 1.000000 -weight_frac 1.0 -maxrot 6 -maxshf 10 -VERB -warp aff -source_automask+4 -twobest 11 -twopass -VERB -maxrot 45 -maxshf 40 -fineblur 1 -source_automask+2
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Option '-cmass' enables center-of-mass code = 7 = +xyz
++ Source dataset: ./__tt_sub-08_T1w_ns+orig.HEAD
++ Base dataset: ./__tt_vr_base_min_outlier_ts_ns+orig.HEAD
++ Loading datasets into memory
++ 1583872 voxels in -source_automask+2
++ largeness ==> set -twobest 29
++ Zero-pad: ybot=8 ytop=3
++ Zero-pad: zbot=7 ztop=5
++ 38906 voxels [15.6%] in weight mask
++ Output dataset ./__tt_sub-08_T1w_ns_al_junk_wtal+orig.BRIK
++ Number of points for matching = 38906
++ NOTE: base and source coordinate systems have different handedness
+ Orientations: base=Right handed (LPI); source=Left handed (RPI)
+ - It is nothing to worry about: 3dAllineate aligns based on coordinates.
+ - But it is always important to check the alignment visually to be sure.
++ Local correlation: blok type = 'TOHD(17.0941)'
++ base center of mass = 31.963 35.048 27.741 (index)
+ source center of mass = 84.800 114.693 116.298 (index)
+ source-target CM = 86.190 -3.441 -1.709 (xyz)
+ estimated center of mass shifts = 86.190 -3.441 -1.709
++ shift param auto-range: 25.5..146.9 -74.7..67.8 -67.2..63.8
+ Range param#4 [z-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#5 [x-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#6 [y-angle] = -6.000000 .. 6.000000 center = 0.000000
+ Range param#1 [x-shift] = 76.190132 .. 96.190132 center = 86.190132
+ Range param#2 [y-shift] = -13.440697 .. 6.559303 center = -3.440697
+ Range param#3 [z-shift] = -11.708786 .. 8.291214 center = -1.708786
+ Range param#4 [z-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#5 [x-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#6 [y-angle] = -45.000000 .. 45.000000 center = 0.000000
+ Range param#1 [x-shift] = 46.190132 .. 126.190132 center = 86.190132
+ Range param#2 [y-shift] = -43.440697 .. 36.559303 center = -3.440697
+ Range param#3 [z-shift] = -41.708786 .. 38.291214 center = -1.708786
+ 12 free parameters
++ Normalized (unitless) convergence radius = 0.0000088
++ Final parameter search ranges:
+ x-shift = 46.190 .. 126.190
+ y-shift = -43.441 .. 36.559
+ z-shift = -41.709 .. 38.291
+ z-angle = -45.000 .. 45.000
+ x-angle = -45.000 .. 45.000
+ y-angle = -45.000 .. 45.000
+ x-scale = 0.711 .. 1.406
+ y-scale = 0.711 .. 1.406
+ z-scale = 0.711 .. 1.406
+ y/x-shear = -0.111 .. 0.111
+ z/x-shear = -0.111 .. 0.111
+ z/y-shear = -0.111 .. 0.111
++ changing output grid spacing to 1.0000 mm
++ OpenMP thread count = 15
++ ======= Allineation of 1 sub-bricks using Local Pearson Correlation Signed =======
+ source mask has 1583872 [out of 11534336] voxels
+ base mask has 52253 [out of 249600] voxels
++ ========== sub-brick #0 ========== [total CPU to here=0.0 s]
++ *** Coarse pass begins ***
+ * Enter alignment setup routine
+ - copying base image
+ - copying source image
+ - Smoothing base; radius=4.00
+ - Smoothing source; radius=4.00
+ !source mask fill: ubot=64 usiz=159.5
+ - copying weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ - Search for coarse starting parameters
+ 32391 total points stored in 63 'TOHD(17.5558)' bloks (0 duplicates)
+ - number of free params = 6
+ - Test (64+191)*64 params [top5=*o+-.]:#o+o-.+.o--.--+oo-.++-.o-+-.+o+o---+.--.oo.+++o+$+.--.+..o.o + - best 88 costs found:
o= 0 v=-0.078395: 86.19 -3.44 -1.71 0.00 0.00 0.00 [grid]
o= 1 v=-0.024170: 91.62 4.15 28.38 -38.19 22.74 -41.67 [rand]
o= 2 v=-0.022830: 100.19 -18.97 33.97 -14.61 -39.00 -28.71 [rand]
o= 3 v=-0.022358: 92.86 -30.11 24.96 7.50 -30.00 -30.00 [grid]
o= 4 v=-0.022101: 121.80 -7.85 -38.58 -37.76 24.22 3.86 [rand]
o= 5 v=-0.021851: 97.68 -19.75 -35.46 -19.17 13.96 2.56 [rand]
o= 6 v=-0.021730: 122.84 -20.67 20.55 5.26 -41.39 -40.03 [rand]
o= 7 v=-0.021515: 101.83 -12.20 -35.54 -23.16 17.17 -3.59 [rand]
o= 8 v=-0.021205: 91.12 -0.47 27.39 -40.99 18.21 -33.12 [rand]
o= 9 v=-0.020806: 121.80 -7.85 -38.58 -37.76 24.22 -3.86 [rand]
o=10 v=-0.020703: 97.68 -19.75 -35.46 -19.17 13.96 -2.56 [rand]
o=11 v=-0.020229: 121.80 0.97 35.16 37.76 -24.22 -3.86 [rand]
o=12 v=-0.020075: 90.34 20.37 30.18 31.52 -11.26 -33.63 [rand]
o=13 v=-0.019908: 100.19 -18.97 33.97 14.61 -39.00 -28.71 [rand]
o=14 v=-0.019694: 120.99 -17.22 -35.72 10.62 -7.10 -10.42 [rand]
o=15 v=-0.019540: 119.22 1.58 -38.52 -11.98 4.28 -19.20 [rand]
o=16 v=-0.019170: 88.48 -10.62 -33.40 -10.97 18.37 5.04 [rand]
o=17 v=-0.018913: 121.80 0.97 -38.58 -37.76 24.22 3.86 [rand]
o=18 v=-0.018894: 96.65 -30.71 19.79 -34.95 -5.03 -42.69 [rand]
o=19 v=-0.018891: 115.81 31.17 35.11 24.31 -11.89 -2.84 [rand]
o=20 v=-0.018703: 92.86 -30.11 24.96 -7.50 -30.00 -30.00 [grid]
o=21 v=-0.018636: 115.81 31.17 35.11 24.31 -11.89 2.84 [rand]
o=22 v=-0.018271: 74.70 12.87 -35.46 19.17 13.96 2.56 [rand]
o=23 v=-0.018249: 83.90 -10.62 -33.40 -10.97 18.37 5.04 [rand]
o=24 v=-0.018182: 119.22 -8.46 -38.52 -11.98 4.28 -19.20 [rand]
o=25 v=-0.018157: 112.86 3.23 -28.38 -30.00 7.50 7.50 [grid]
o=26 v=-0.018092: 113.03 -16.54 -34.29 -39.07 28.37 8.01 [rand]
o=27 v=-0.018024: 121.80 -7.85 35.16 37.76 -24.22 -3.86 [rand]
o=28 v=-0.017957: 91.12 -6.41 27.39 -40.99 18.21 -33.12 [rand]
o=29 v=-0.017956: 101.83 -12.20 -35.54 -23.16 17.17 3.59 [rand]
o=30 v=-0.017508: 120.99 -17.22 -35.72 -10.62 -7.10 -10.42 [rand]
o=31 v=-0.017362: 74.70 12.87 -35.46 19.17 13.96 -2.56 [rand]
o=32 v=-0.017349: 120.99 -17.22 -35.72 -10.62 7.10 -10.42 [rand]
o=33 v=-0.016693: 105.31 19.97 -35.01 -37.88 13.88 18.42 [rand]
o=34 v=-0.016675: 102.99 15.02 30.65 10.10 -16.32 -39.31 [rand]
o=35 v=-0.016669: 105.31 19.97 -35.01 -37.88 13.88 -18.42 [rand]
o=36 v=-0.016632: 120.99 -17.22 -35.72 10.62 7.10 -10.42 [rand]
o=37 v=-0.016450: 99.53 -27.69 29.08 6.84 -32.76 -20.90 [rand]
o=38 v=-0.016261: 79.52 -30.11 24.96 -30.00 -7.50 -30.00 [grid]
o=39 v=-0.016134: 99.35 34.44 35.49 14.03 -8.39 -25.20 [rand]
o=40 v=-0.016094: 79.78 30.50 -38.28 -19.28 27.13 15.13 [rand]
o=41 v=-0.015858: 95.39 -15.21 -23.44 -6.82 34.51 24.39 [rand]
o=42 v=-0.015801: 120.99 -17.22 -35.72 10.62 -7.10 10.42 [rand]
o=43 v=-0.015734: 70.55 5.32 -35.54 23.16 17.17 -3.59 [rand]
o=44 v=-0.015723: 67.07 19.97 -35.01 37.88 13.88 -18.42 [rand]
o=45 v=-0.015700: 92.86 23.23 24.96 30.00 -7.50 -30.00 [grid]
o=46 v=-0.015623: 113.12 -31.58 -39.65 15.05 4.71 -12.78 [rand]
o=47 v=-0.015567: 92.86 -10.11 -28.38 -7.50 7.50 30.00 [grid]
o=48 v=-0.015330: 88.48 -10.62 -33.40 -10.97 18.37 -5.04 [rand]
o=49 v=-0.015192: 92.03 -32.53 20.97 30.43 -41.57 -36.66 [rand]
o=50 v=-0.015111: 103.24 -26.13 22.54 -12.15 -12.44 -39.75 [rand]
o=51 v=-0.015075: 122.78 -30.74 29.53 -25.97 -37.53 -40.21 [rand]
o=52 v=-0.014925: 92.86 -10.11 -28.38 -7.50 30.00 30.00 [grid]
o=53 v=-0.014904: 121.80 0.97 -38.58 -37.76 24.22 -3.86 [rand]
o=54 v=-0.014835: 102.99 -21.90 30.65 -10.10 -16.32 -39.31 [rand]
o=55 v=-0.014711: 102.99 15.02 -34.07 -10.10 -16.32 39.31 [rand]
o=56 v=-0.014651: 119.22 -8.46 -38.52 -11.98 -4.28 -19.20 [rand]
o=57 v=-0.014527: 99.35 -41.33 -38.90 14.03 8.39 -25.20 [rand]
o=58 v=-0.014469: 70.55 -12.20 -35.54 23.16 17.17 3.59 [rand]
o=59 v=-0.014214: 121.80 -7.85 35.16 37.76 -24.22 3.86 [rand]
o=60 v=-0.014171: 70.55 -12.20 -35.54 23.16 17.17 -3.59 [rand]
o=61 v=-0.014132: 94.85 -27.60 34.86 27.41 -42.10 -24.35 [rand]
o=62 v=-0.014108: 92.86 -10.11 -28.38 30.00 7.50 7.50 [grid]
o=63 v=-0.013985: 92.61 -37.38 -38.28 19.28 27.13 -15.13 [rand]
o=64 v=-0.013771: 56.91 -19.20 28.15 13.45 -19.96 14.96 [rand]
o=65 v=-0.013738: 104.93 -39.84 -34.76 24.90 4.74 -17.23 [rand]
o=66 v=-0.013600: 92.86 3.23 -28.38 -7.50 30.00 30.00 [grid]
o=67 v=-0.013545: 102.99 15.02 -34.07 10.10 -16.32 39.31 [rand]
o=68 v=-0.013506: 97.68 12.87 -35.46 -19.17 13.96 -2.56 [rand]
o=69 v=-0.013420: 92.86 3.23 24.96 30.00 -30.00 -30.00 [grid]
o=70 v=-0.013384: 83.90 -10.62 -33.40 -10.97 18.37 -5.04 [rand]
o=71 v=-0.013341: 115.81 -38.05 -38.52 24.31 -11.89 -2.84 [rand]
o=72 v=-0.013295: 59.26 -31.58 36.24 15.05 -4.71 12.78 [rand]
o=73 v=-0.013221: 79.78 30.50 -38.28 19.28 27.13 15.13 [rand]
o=74 v=-0.013191: 59.35 9.66 -34.29 39.07 28.37 -8.01 [rand]
o=75 v=-0.013149: 118.40 -32.49 34.43 -40.41 -19.09 -40.75 [rand]
o=76 v=-0.013072: 70.55 5.32 -35.54 23.16 17.17 3.59 [rand]
o=77 v=-0.013043: 65.94 23.84 -36.19 37.90 5.03 3.08 [rand]
o=78 v=-0.012929: 92.86 -10.11 -28.38 7.50 7.50 7.50 [grid]
o=79 v=-0.012857: 60.67 -34.78 32.57 42.46 -41.07 35.63 [rand]
o=80 v=-0.012789: 99.35 -41.33 35.49 14.03 -8.39 -25.20 [rand]
o=81 v=-0.012665: 97.68 12.87 -35.46 -19.17 13.96 2.56 [rand]
o=82 v=-0.012652: 112.86 -30.11 -28.38 30.00 7.50 7.50 [grid]
o=83 v=-0.012596: 104.93 -39.84 31.34 -24.90 -4.74 -17.23 [rand]
o=84 v=-0.012570: 83.90 -10.62 -33.40 10.97 18.37 -5.04 [rand]
o=85 v=-0.012553: 92.86 -10.11 -28.38 -7.50 30.00 7.50 [grid]
o=86 v=-0.012529: 92.86 -10.11 24.96 -7.50 -7.50 -30.00 [grid]
o=87 v=-0.012519: 80.37 10.75 27.93 38.21 -15.41 -13.51 [rand]
+ - A little optimization:*[#16335=-0.0833147] *[#16338=-0.0996876] *[#16339=-0.122258] *[#16342=-0.125257] *[#16344=-0.126031] *[#16346=-0.127847] *[#16349=-0.129014] *[#16352=-0.129155] *[#16355=-0.129328] *[#16356=-0.129367] *[#16360=-0.12939] *[#16362=-0.129425] *[#16363=-0.129437] *[#16366=-0.129478] ........................................................................................
+ - costs of the above after a little optimization:
*o= 0 v=-0.129478: 86.56 -2.92 0.24 -1.17 0.39 0.22 [grid] [f=50]
o= 1 v=-0.025208: 91.85 3.62 28.37 -37.98 22.65 -41.84 [rand] [f=41]
o= 2 v=-0.037638: 97.07 -22.70 36.86 -10.25 -41.88 -25.72 [rand] [f=62]
o= 3 v=-0.026228: 94.01 -30.11 26.00 6.89 -35.26 -29.95 [grid] [f=67]
o= 4 v=-0.025109: 122.87 -3.88 -38.32 -37.64 24.31 3.06 [rand] [f=61]
o= 5 v=-0.023467: 97.59 -19.20 -35.11 -18.55 14.45 2.78 [rand] [f=47]
o= 6 v=-0.032558: 124.34 -28.56 20.34 5.63 -41.94 -44.51 [rand] [f=52]
o= 7 v=-0.024558: 103.87 -12.91 -36.21 -26.12 14.64 -5.56 [rand] [f=68]
o= 8 v=-0.026595: 92.49 0.17 27.38 -37.57 16.88 -34.60 [rand] [f=47]
o= 9 v=-0.025748: 122.87 -7.62 -36.96 -38.73 25.59 5.74 [rand] [f=37]
o=10 v=-0.023050: 97.81 -19.65 -34.79 -19.15 13.89 2.05 [rand] [f=36]
o=11 v=-0.034817: 125.56 3.51 37.77 32.42 -27.27 -8.86 [rand] [f=47]
o=12 v=-0.026636: 90.62 19.43 30.46 30.75 -11.47 -27.04 [rand] [f=54]
o=13 v=-0.029615: 95.65 -16.42 33.99 15.06 -38.94 -27.36 [rand] [f=44]
o=14 v=-0.025724: 121.05 -17.94 -34.73 10.44 -8.00 -4.28 [rand] [f=40]
o=15 v=-0.027042: 119.10 0.10 -39.89 -13.81 11.33 -14.01 [rand] [f=42]
o=16 v=-0.029803: 88.38 -15.30 -33.36 -10.38 12.73 4.37 [rand] [f=58]
o=17 v=-0.025471: 120.77 -7.35 -37.30 -34.55 25.97 6.34 [rand] [f=51]
o=18 v=-0.021813: 96.06 -31.35 20.30 -37.16 -7.69 -44.56 [rand] [f=37]
o=19 v=-0.039610: 120.38 31.62 37.96 31.21 -11.89 -1.49 [rand] [f=60]
o=20 v=-0.030208: 99.98 -30.12 26.92 -4.50 -28.25 -34.17 [grid] [f=62]
o=21 v=-0.033973: 115.95 30.51 37.09 27.08 -7.23 4.45 [rand] [f=45]
o=22 v=-0.024448: 68.66 12.44 -38.44 19.11 14.37 2.63 [rand] [f=52]
o=23 v=-0.030816: 88.36 -12.05 -34.28 -12.31 11.56 5.36 [rand] [f=57]
o=24 v=-0.026831: 119.70 -7.09 -37.52 -10.99 4.50 -13.47 [rand] [f=43]
o=25 v=-0.021834: 110.54 4.11 -28.89 -31.19 6.72 7.75 [grid] [f=46]
o=26 v=-0.029659: 112.69 -13.96 -35.31 -28.09 23.99 4.39 [rand] [f=64]
o=27 v=-0.036380: 124.25 -11.62 38.17 33.28 -31.14 -4.50 [rand] [f=57]
o=28 v=-0.026330: 92.18 1.00 27.77 -39.33 18.23 -32.61 [rand] [f=51]
o=29 v=-0.022613: 102.63 -11.90 -37.10 -25.08 13.86 -0.27 [rand] [f=35]
o=30 v=-0.023502: 119.40 -16.34 -38.41 -9.52 -2.70 -13.24 [rand] [f=49]
o=31 v=-0.024601: 69.46 14.02 -37.34 19.44 12.69 -2.24 [rand] [f=44]
o=32 v=-0.025824: 119.43 -8.11 -37.98 -7.01 10.78 -13.52 [rand] [f=83]
o=33 v=-0.028485: 111.32 17.78 -33.51 -44.98 10.27 21.60 [rand] [f=50]
o=34 v=-0.021606: 103.05 11.34 30.91 10.44 -16.63 -39.34 [rand] [f=54]
o=35 v=-0.025812: 105.92 18.80 -34.03 -38.48 13.27 -12.88 [rand] [f=49]
o=36 v=-0.023639: 120.56 -21.02 -38.02 5.46 4.63 -15.69 [rand] [f=51]
o=37 v=-0.022421: 96.59 -28.00 29.09 6.71 -31.63 -25.55 [rand] [f=39]
o=38 v=-0.020190: 79.59 -26.31 24.88 -29.92 -8.68 -28.78 [grid] [f=49]
o=39 v=-0.021749: 96.94 33.57 35.86 11.35 -6.94 -19.87 [rand] [f=66]
o=40 v=-0.019895: 82.95 28.96 -37.93 -17.75 28.76 13.02 [rand] [f=37]
o=41 v=-0.021356: 91.66 -14.96 -22.76 -7.10 34.58 24.82 [rand] [f=46]
o=42 v=-0.027107: 124.49 -20.18 -32.05 10.69 -7.44 9.91 [rand] [f=45]
o=43 v=-0.026319: 68.91 11.17 -38.05 20.29 19.40 -6.43 [rand] [f=56]
o=44 v=-0.024913: 67.84 15.42 -35.49 37.39 14.37 -18.96 [rand] [f=37]
o=45 v=-0.023354: 93.76 23.30 27.77 29.47 -7.75 -31.27 [grid] [f=40]
o=46 v=-0.019407: 109.48 -31.82 -39.14 15.51 4.62 -13.43 [rand] [f=46]
o=47 v=-0.022879: 92.79 -14.50 -30.57 -6.67 8.67 35.86 [grid] [f=59]
o=48 v=-0.028698: 88.61 -13.59 -36.48 -10.26 12.62 0.84 [rand] [f=53]
o=49 v=-0.019705: 92.37 -28.32 21.44 30.84 -39.72 -37.84 [rand] [f=65]
o=50 v=-0.022741: 99.40 -24.95 21.80 -13.00 -13.24 -40.83 [rand] [f=46]
o=51 v=-0.025023: 125.97 -26.96 37.93 -27.17 -36.78 -38.74 [rand] [f=57]
o=52 v=-0.018040: 92.91 -9.23 -27.61 -8.65 30.90 29.96 [grid] [f=65]
o=53 v=-0.026083: 119.13 -8.06 -39.09 -31.95 24.41 -0.92 [rand] [f=47]
o=54 v=-0.021481: 97.21 -15.76 28.19 -9.68 -14.20 -39.20 [rand] [f=52]
o=55 v=-0.029622: 102.22 23.54 -40.40 -8.51 -15.66 40.94 [rand] [f=57]
o=56 v=-0.024318: 119.26 -12.01 -40.99 -6.33 -3.92 -15.68 [rand] [f=67]
o=57 v=-0.016309: 99.90 -40.50 -41.08 13.83 7.03 -28.14 [rand] [f=41]
o=58 v=-0.020067: 70.82 -7.68 -36.96 23.60 18.14 3.28 [rand] [f=53]
o=59 v=-0.029083: 122.02 -8.54 38.29 35.74 -24.75 2.19 [rand] [f=40]
o=60 v=-0.018760: 69.81 -16.27 -37.23 22.97 16.08 -3.59 [rand] [f=33]
o=61 v=-0.027637: 95.70 -14.89 33.59 24.68 -37.62 -25.78 [rand] [f=55]
o=62 v=-0.021869: 90.87 -7.14 -30.34 27.19 5.03 7.77 [grid] [f=44]
o=63 v=-0.016562: 93.04 -33.28 -38.26 19.24 27.71 -15.44 [rand] [f=35]
o=64 v=-0.017593: 53.52 -19.20 30.34 13.28 -18.83 16.03 [rand] [f=60]
o=65 v=-0.020603: 105.13 -39.25 -35.37 22.69 13.32 -21.57 [rand] [f=50]
o=66 v=-0.022279: 93.18 9.76 -30.87 -8.28 32.53 27.85 [grid] [f=76]
o=67 v=-0.023893: 102.80 20.21 -36.92 8.62 -14.85 40.53 [rand] [f=50]
o=68 v=-0.030230: 94.78 11.44 -41.70 -27.01 18.02 -15.08 [rand] [f=53]
o=69 v=-0.025604: 92.34 2.49 30.25 29.16 -28.24 -27.67 [grid] [f=63]
o=70 v=-0.028099: 84.44 -14.51 -37.03 -5.85 10.74 5.83 [rand] [f=62]
o=71 v=-0.024499: 117.76 -30.97 -33.13 30.69 -12.85 -3.72 [rand] [f=61]
o=72 v=-0.017104: 59.48 -35.24 36.02 15.19 -4.00 12.18 [rand] [f=44]
o=73 v=-0.020121: 78.49 34.05 -38.37 19.48 26.09 14.47 [rand] [f=43]
o=74 v=-0.027725: 66.37 14.95 -34.10 33.91 20.53 -10.37 [rand] [f=70]
o=75 v=-0.021457: 118.15 -34.24 32.27 -35.98 -26.19 -42.12 [rand] [f=53]
o=76 v=-0.033583: 84.74 5.08 -40.21 22.43 10.74 3.93 [rand] [f=82]
o=77 v=-0.022471: 66.16 19.67 -38.88 38.58 4.87 -0.04 [rand] [f=62]
o=78 v=-0.022165: 93.72 -7.71 -26.28 17.15 7.89 8.13 [grid] [f=39]
o=79 v=-0.019392: 56.80 -35.73 32.75 42.93 -41.01 35.05 [rand] [f=40]
o=80 v=-0.024286: 99.09 -41.34 36.23 14.84 -11.57 -20.31 [rand] [f=40]
o=81 v=-0.027989: 100.32 13.71 -36.07 -26.64 6.16 -4.67 [rand] [f=61]
o=82 v=-0.018578: 114.04 -25.07 -27.78 31.22 5.83 3.98 [grid] [f=41]
o=83 v=-0.020005: 108.98 -40.26 31.30 -22.78 -4.31 -16.42 [rand] [f=52]
o=84 v=-0.022922: 85.64 -10.65 -38.37 5.12 14.67 -7.14 [rand] [f=60]
o=85 v=-0.022479: 89.13 -11.37 -28.03 -5.87 37.76 7.84 [grid] [f=59]
o=86 v=-0.019822: 93.58 -9.73 28.66 -9.65 -11.27 -31.86 [grid] [f=57]
o=87 v=-0.031659: 80.56 10.85 31.85 39.61 -23.66 -12.54 [rand] [f=68]
+ - saving # 0 for use with twobest
+ - saving #19 for use with twobest
+ - saving # 2 for use with twobest
+ - saving #27 for use with twobest
+ - saving #11 for use with twobest
+ - saving #21 for use with twobest
+ - saving #76 for use with twobest
+ - saving # 6 for use with twobest
+ - saving #87 for use with twobest
+ - saving #23 for use with twobest
+ - saving #68 for use with twobest
+ - saving #20 for use with twobest
+ - skip #16 for twobest: too close to set #23
+ - saving #26 for use with twobest
+ - saving #55 for use with twobest
+ - saving #13 for use with twobest
+ - saving #59 for use with twobest
+ - skip #48 for twobest: too close to set #16
+ - saving #33 for use with twobest
+ - saving #70 for use with twobest
+ - saving #81 for use with twobest
+ - saving #74 for use with twobest
+ - saving #61 for use with twobest
+ - saving #42 for use with twobest
+ - saving #15 for use with twobest
+ - saving #24 for use with twobest
+ - saving #12 for use with twobest
+ - saving # 8 for use with twobest
+ - skip #28 for twobest: too close to set # 8
+ - saving #43 for use with twobest
+ - saving # 3 for use with twobest
+ - saving #53 for use with twobest
+ - Coarse startup search net CPU time = 0.0 s
++ Start refinement #1 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=3.11
+ - Smoothing source; radius=3.11
+ !source mask fill: ubot=64 usiz=159.5
+ - retaining old weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31909 total points stored in 63 'TOHD(17.3748)' bloks (0 duplicates)
*[#20978=-0.169331] *[#20986=-0.170313] *[#21000=-0.170526] *[#21001=-0.172023] *[#21005=-0.172274] *[#21008=-0.173681] *[#21009=-0.1739] *[#21013=-0.174339] *[#21016=-0.175108] *[#21017=-0.176175] *[#21018=-0.176452] *[#21019=-0.176809] *[#21021=-0.177261] *[#21024=-0.17761] *[#21026=-0.178186] *[#21027=-0.178833] *[#21028=-0.179343] *[#21031=-0.179472] *[#21040=-0.180395] *[#21043=-0.180428] *[#21045=-0.180429] *[#21047=-0.180514] *[#21048=-0.180599] *[#21049=-0.180672] *[#21051=-0.180752] *[#21052=-0.180786] *[#21053=-0.180808] *[#21054=-0.18082] *[#21055=-0.180832] *[#21056=-0.180858] *[#21057=-0.180864] *[#21059=-0.180909] *[#21060=-0.180915] *[#21061=-0.180934] *[#21062=-0.181065] *[#21063=-0.181087] *[#21064=-0.181114] *[#21065=-0.181253] *[#21068=-0.181324] *[#21069=-0.181432] *[#21070=-0.18147] *[#21073=-0.181504] *[#21074=-0.18155] *[#21075=-0.181565]
+ - param set #1 has cost=-0.181565 [o=0 t=0]
+ -- Parameters = 86.2417 -3.0340 0.1839 -0.7927 0.0198 0.1572 0.9849 1.0186 0.9937 -0.0067 -0.0009 -0.0030
+ - param set #2 has cost=-0.029599 [o=19 t=1]
+ -- Parameters = 120.3747 31.4269 37.8606 31.0260 -11.8449 -1.1094 1.0067 0.9979 0.9877 -0.0009 0.0021 -0.0001
+ - param set #3 has cost=-0.032947 [o=2 t=2]
+ -- Parameters = 97.1422 -22.9068 36.7125 -10.4271 -42.0244 -25.8414 0.9953 0.9994 0.9958 -0.0016 0.0013 -0.0002
+ - param set #4 has cost=-0.025913 [o=27 t=3]
+ -- Parameters = 124.2213 -11.7386 37.9231 33.3757 -31.0928 -4.5253 1.0015 1.0012 0.9970 0.0002 0.0015 0.0004
+ - param set #5 has cost=-0.028112 [o=11 t=4]
+ -- Parameters = 122.3987 3.2658 37.8554 32.0364 -27.2812 -8.8197 0.9993 1.0000 0.9989 -0.0002 -0.0001 -0.0002
+ - param set #6 has cost=-0.027098 [o=21 t=5]
+ -- Parameters = 115.6655 30.6513 37.9152 27.7891 -10.1989 1.4674 0.9984 0.9912 0.9884 0.0004 -0.0012 0.0008
+ - param set #7 has cost=-0.025330 [o=76 t=6]
+ -- Parameters = 84.5296 4.8670 -40.1157 22.4018 10.4986 3.9660 1.0038 0.9999 1.0018 -0.0005 -0.0003 0.0016
+ - param set #8 has cost=-0.027671 [o=6 t=7]
+ -- Parameters = 124.4184 -28.3909 20.3705 5.2792 -41.8702 -44.1177 1.0047 0.9790 1.0062 -0.0035 -0.0002 -0.0054
+ - param set #9 has cost=-0.026880 [o=87 t=8]
+ -- Parameters = 80.6211 10.7956 31.8627 39.3826 -23.6462 -12.7049 0.9977 1.0051 0.9899 -0.0002 0.0006 -0.0006
+ - param set #10 has cost=-0.025438 [o=23 t=9]
+ -- Parameters = 88.5119 -12.3057 -34.5255 -12.2625 11.3388 4.8335 1.0036 0.9951 0.9911 0.0147 -0.0003 -0.0010
+ - param set #11 has cost=-0.025056 [o=68 t=10]
+ -- Parameters = 95.0321 11.2848 -41.6982 -27.1384 18.0539 -14.7625 0.9967 0.9975 1.0046 0.0010 -0.0017 -0.0002
+ - param set #12 has cost=-0.026301 [o=20 t=11]
+ -- Parameters = 99.7295 -29.7606 26.8107 -4.8322 -28.5727 -34.7626 1.0025 0.9939 1.0000 0.0001 -0.0010 0.0011
+ - param set #13 has cost=-0.024785 [o=26 t=12]
+ -- Parameters = 112.7484 -13.9672 -35.1899 -28.0512 24.1536 4.0959 0.9974 1.0021 1.0031 -0.0009 0.0004 -0.0004
+ - param set #14 has cost=-0.023818 [o=55 t=13]
+ -- Parameters = 102.3059 23.5118 -40.3609 -8.3749 -15.9182 41.1731 0.9987 1.0004 1.0059 0.0001 -0.0012 -0.0005
+ - param set #15 has cost=-0.025346 [o=13 t=14]
+ -- Parameters = 95.5049 -16.5664 34.3042 14.7561 -38.9196 -27.3528 1.0042 1.0061 0.9993 0.0005 -0.0002 -0.0014
+ - param set #16 has cost=-0.028890 [o=59 t=15]
+ -- Parameters = 121.0495 -8.3479 38.2577 35.4740 -28.4600 -2.1114 0.9475 0.9673 0.9742 0.0001 -0.0099 0.0125
+ - param set #17 has cost=-0.022857 [o=33 t=16]
+ -- Parameters = 111.2125 17.7604 -33.8452 -44.8026 10.9680 21.5318 0.9972 1.0001 1.0062 -0.0000 0.0110 -0.0001
+ - param set #18 has cost=-0.024121 [o=70 t=17]
+ -- Parameters = 84.6139 -14.8654 -35.5807 -6.2385 9.4374 4.5597 0.9971 1.0019 0.9957 -0.0008 0.0001 -0.0020
+ - param set #19 has cost=-0.026862 [o=81 t=18]
+ -- Parameters = 100.4019 15.0228 -35.6483 -31.5537 6.5158 -4.5506 0.9870 0.9952 0.9971 0.0007 0.0006 0.0008
+ - param set #20 has cost=-0.019718 [o=74 t=19]
+ -- Parameters = 66.3511 14.9204 -34.0041 33.9302 20.6377 -10.4785 0.9995 1.0003 0.9990 0.0002 -0.0001 -0.0002
+ - param set #21 has cost=-0.024596 [o=61 t=20]
+ -- Parameters = 94.9530 -15.7971 34.3648 21.1914 -38.2038 -26.5993 0.9967 1.0153 0.9925 0.0030 0.0004 -0.0029
+ - param set #22 has cost=-0.030211 [o=42 t=21]
+ -- Parameters = 125.4314 -20.5678 -30.0616 11.1232 -9.1532 7.8234 0.9842 0.9823 0.9478 -0.0144 -0.0209 0.0117
+ - param set #23 has cost=-0.022798 [o=15 t=22]
+ -- Parameters = 119.2795 0.0863 -39.9379 -13.8465 11.2937 -14.2121 0.9972 0.9982 1.0023 0.0015 -0.0005 -0.0005
+ - param set #24 has cost=-0.022617 [o=24 t=23]
+ -- Parameters = 119.5177 -7.0698 -37.4677 -10.8037 4.7487 -13.3916 1.0004 1.0044 0.9954 0.0003 -0.0039 -0.0012
+ - param set #25 has cost=-0.025054 [o=12 t=24]
+ -- Parameters = 90.2415 19.2854 31.2812 30.8214 -12.2280 -26.6292 0.9929 0.9856 0.9532 -0.0054 0.0058 -0.0182
+ - param set #26 has cost=-0.022589 [o=8 t=25]
+ -- Parameters = 92.2565 -0.3541 27.7222 -37.8739 16.7082 -34.1457 1.0015 0.9972 0.9965 0.0001 0.0001 -0.0009
+ - param set #27 has cost=-0.023658 [o=43 t=26]
+ -- Parameters = 67.8001 10.6026 -38.1869 20.2024 17.6918 -6.5006 0.9989 0.9962 0.9715 0.0005 -0.0034 0.0218
+ - param set #28 has cost=-0.023512 [o=3 t=27]
+ -- Parameters = 93.6271 -30.3348 25.2672 6.4093 -34.2989 -30.7456 1.0028 0.9979 1.0102 0.0018 0.0004 -0.0028
+ - param set #29 has cost=-0.022179 [o=53 t=28]
+ -- Parameters = 119.1053 -8.1075 -39.0073 -32.2735 24.1427 -0.5844 0.9994 1.0016 1.0011 -0.0012 0.0000 0.0001
*[#23803=-0.181994] *[#23804=-0.182059] *[#23808=-0.182443] *[#23813=-0.182802] *[#23817=-0.182824] *[#23821=-0.183125]
+ - param set #30 has cost=-0.183125 [o=-1 t=-1]
+ -- Parameters = 86.2790 -2.9386 0.2322 -0.5467 -0.0139 0.1276 0.9790 1.0183 0.9889 0.0075 0.0058 -0.0075
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0636
+ --- dist(#3,#1) = 0.467
+ --- dist(#4,#1) = 0.489
+ --- dist(#5,#1) = 0.47
+ --- dist(#6,#1) = 0.475
+ --- dist(#7,#1) = 0.47
+ --- dist(#8,#1) = 0.492
+ --- dist(#9,#1) = 0.471
+ --- dist(#10,#1) = 0.444
+ --- dist(#11,#1) = 0.449
+ --- dist(#12,#1) = 0.388
+ --- dist(#13,#1) = 0.474
+ --- dist(#14,#1) = 0.434
+ --- dist(#15,#1) = 0.432
+ --- dist(#16,#1) = 0.504
+ --- dist(#17,#1) = 0.524
+ --- dist(#18,#1) = 0.388
+ --- dist(#19,#1) = 0.443
+ --- dist(#20,#1) = 0.427
+ --- dist(#21,#1) = 0.448
+ --- dist(#22,#1) = 0.507
+ --- dist(#23,#1) = 0.48
+ --- dist(#24,#1) = 0.381
+ --- dist(#25,#1) = 0.492
+ --- dist(#26,#1) = 0.502
+ --- dist(#27,#1) = 0.471
+ --- dist(#28,#1) = 0.415
+ --- dist(#29,#1) = 0.49
+ --- dist(#30,#1) = 0.428
++ Start refinement #2 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=2.42
+ - Smoothing source; radius=2.42
+ !source mask fill: ubot=64 usiz=159.5
+ - retaining old weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 31999 total points stored in 64 'TOHD(17.2644)' bloks (0 duplicates)
*[#23823=-0.208583] *[#23853=-0.209083] *[#23856=-0.209692] *[#23857=-0.21004] *[#23858=-0.210473] *[#23859=-0.210652] *[#23861=-0.210912] *[#23862=-0.21108] *[#23863=-0.211096] *[#23864=-0.211318] *[#23865=-0.211465] *[#23868=-0.211558] *[#23871=-0.211598] *[#23874=-0.211605] *[#23876=-0.211804] *[#23880=-0.211892] *[#23883=-0.212487] *[#23884=-0.212638] *[#23887=-0.212807] *[#23889=-0.212858] *[#23890=-0.212946] *[#23891=-0.213045] *[#23892=-0.213054] *[#23893=-0.213085] *[#23896=-0.213618] *[#23897=-0.214048] *[#23898=-0.2143] *[#23899=-0.214697] *[#23902=-0.214782] *[#23904=-0.215131] *[#23907=-0.215508] *[#23908=-0.21602] *[#23909=-0.21655] *[#23910=-0.216938] *[#23911=-0.21729] *[#23912=-0.217316] *[#23913=-0.217499] *[#23914=-0.217762] *[#23915=-0.217858] *[#23920=-0.218401] *[#23921=-0.218462] *[#23922=-0.218657] *[#23925=-0.219463] *[#23926=-0.219544] *[#23927=-0.219959] *[#23928=-0.220151] *[#23931=-0.220227]
+ - param set #1 has cost=-0.220227 [o=-1 t=-1]
+ -- Parameters = 86.5690 -3.1646 -0.1381 -0.8542 -1.9133 -0.7763 0.9848 1.0171 0.9535 -0.0108 0.0209 -0.0382
+ - param set #2 has cost=-0.216445 [o=0 t=0]
+ -- Parameters = 86.4148 -3.1639 -0.1179 -0.8652 -0.7590 -0.0405 0.9867 1.0242 0.9680 -0.0151 0.0021 -0.0159
+ - param set #3 has cost=-0.027810 [o=2 t=2]
+ -- Parameters = 97.2032 -22.9052 36.6921 -10.4554 -42.1013 -25.7685 0.9914 0.9972 0.9931 -0.0021 0.0004 0.0019
+ - param set #4 has cost=-0.030534 [o=42 t=21]
+ -- Parameters = 124.9320 -22.0031 -26.3056 8.7231 -2.9528 10.9929 0.9078 0.9429 0.9127 -0.0312 -0.0436 -0.0116
+ - param set #5 has cost=-0.024661 [o=19 t=1]
+ -- Parameters = 120.2512 31.5807 37.8833 31.4755 -11.7813 -0.9877 1.0280 0.9922 0.9858 -0.0015 0.0013 0.0011
+ - param set #6 has cost=-0.022586 [o=59 t=15]
+ -- Parameters = 121.0138 -8.5765 38.2813 35.5698 -28.5095 -2.1222 0.9444 0.9668 0.9787 0.0009 -0.0099 0.0127
+ - param set #7 has cost=-0.024438 [o=11 t=4]
+ -- Parameters = 122.3828 3.2625 37.8905 32.0936 -27.2969 -8.9957 0.9996 1.0008 0.9996 -0.0003 -0.0004 0.0002
+ - param set #8 has cost=-0.022973 [o=6 t=7]
+ -- Parameters = 124.5990 -28.5221 20.3574 5.1992 -41.4910 -44.5459 1.0133 0.9665 0.9986 -0.0044 -0.0021 -0.0082
+ - param set #9 has cost=-0.023141 [o=21 t=5]
+ -- Parameters = 115.3082 31.2509 37.9653 28.2268 -10.2700 2.1299 0.9881 0.9725 1.0137 0.0031 -0.0002 -0.0013
+ - param set #10 has cost=-0.020712 [o=87 t=8]
+ -- Parameters = 80.6871 10.7820 31.8608 39.2432 -23.8256 -12.8107 0.9960 1.0010 0.9862 0.0001 -0.0012 -0.0004
+ - param set #11 has cost=-0.023867 [o=81 t=18]
+ -- Parameters = 100.9144 15.5630 -35.5687 -30.9887 7.2817 -4.8546 0.9920 1.0234 0.9953 -0.0013 0.0007 -0.0033
+ - param set #12 has cost=-0.022773 [o=20 t=11]
+ -- Parameters = 99.3746 -29.8610 26.9858 -5.3618 -28.5046 -34.3145 1.0066 0.9859 1.0136 -0.0014 0.0000 0.0010
+ - param set #13 has cost=-0.021875 [o=27 t=3]
+ -- Parameters = 124.5604 -15.0908 37.9811 33.1165 -31.3178 -4.2711 1.0069 1.0029 0.9948 0.0004 0.0007 0.0018
+ - param set #14 has cost=-0.018972 [o=23 t=9]
+ -- Parameters = 88.7944 -12.1116 -34.5232 -12.0411 11.5798 4.8169 1.0017 0.9955 0.9899 0.0150 0.0007 -0.0000
+ - param set #15 has cost=-0.022093 [o=13 t=14]
+ -- Parameters = 94.6722 -16.0639 34.9181 15.7936 -38.3548 -28.1825 0.9908 0.9977 0.9904 -0.0019 -0.0004 -0.0028
+ - param set #16 has cost=-0.019604 [o=76 t=6]
+ -- Parameters = 84.2874 4.5998 -40.1267 21.8844 10.7009 4.2715 1.0055 0.9996 1.0026 -0.0011 -0.0010 0.0040
+ - param set #17 has cost=-0.018545 [o=68 t=10]
+ -- Parameters = 94.9844 11.4663 -41.6971 -27.1688 18.1245 -14.7945 0.9941 1.0035 1.0045 0.0090 -0.0015 -0.0008
+ - param set #18 has cost=-0.022035 [o=12 t=24]
+ -- Parameters = 90.4553 19.1335 31.0461 30.9460 -12.3561 -27.0333 0.9931 0.9841 0.9491 -0.0063 0.0070 -0.0183
+ - param set #19 has cost=-0.017809 [o=26 t=12]
+ -- Parameters = 112.7930 -14.0675 -34.9825 -28.0190 24.1066 3.9824 0.9955 1.0005 1.0055 -0.0008 0.0082 -0.0007
+ - param set #20 has cost=-0.022233 [o=61 t=20]
+ -- Parameters = 94.9835 -15.7922 34.2528 21.3591 -38.3023 -26.6743 0.9942 1.0155 0.9949 0.0025 0.0007 -0.0032
+ - param set #21 has cost=-0.021028 [o=70 t=17]
+ -- Parameters = 86.4718 -14.4547 -34.6323 -6.2583 9.6649 3.7010 0.9942 0.9844 0.9884 0.0090 0.0005 -0.0028
+ - param set #22 has cost=-0.017872 [o=55 t=13]
+ -- Parameters = 102.4708 23.5267 -40.2705 -8.5359 -16.0124 41.0981 1.0009 0.9972 1.0084 -0.0001 -0.0017 -0.0010
+ - param set #23 has cost=-0.017721 [o=43 t=26]
+ -- Parameters = 67.4275 10.6734 -38.2748 20.0915 17.9131 -6.3259 0.9971 0.9935 0.9725 0.0008 0.0052 0.0220
+ - param set #24 has cost=-0.020349 [o=3 t=27]
+ -- Parameters = 92.5580 -30.5120 25.0446 5.9667 -33.9224 -32.3783 0.9901 0.9827 0.9962 0.0134 0.0054 -0.0044
+ - param set #25 has cost=-0.018272 [o=33 t=16]
+ -- Parameters = 110.7879 17.5217 -34.1652 -44.9186 11.3923 22.6426 0.9815 0.9941 0.9763 -0.0011 0.0224 -0.0024
+ - param set #26 has cost=-0.016165 [o=15 t=22]
+ -- Parameters = 119.3371 0.1366 -40.0015 -13.9032 11.2916 -14.2405 0.9960 0.9969 1.0033 0.0023 -0.0007 -0.0005
+ - param set #27 has cost=-0.017790 [o=24 t=23]
+ -- Parameters = 119.5484 -7.0770 -37.5706 -10.9167 4.8044 -13.1528 1.0001 1.0034 0.9886 -0.0026 0.0023 -0.0026
+ - param set #28 has cost=-0.021054 [o=8 t=25]
+ -- Parameters = 92.1249 -0.7941 28.1167 -37.9315 16.4735 -34.0354 0.9818 0.9814 0.9791 -0.0018 -0.0013 -0.0009
+ - param set #29 has cost=-0.015293 [o=53 t=28]
+ -- Parameters = 119.0922 -8.1040 -38.9836 -32.2314 24.1018 -0.5725 1.0000 1.0027 1.0031 -0.0012 0.0001 0.0002
+ - param set #30 has cost=-0.014662 [o=74 t=19]
+ -- Parameters = 66.4003 14.9545 -34.0002 33.9259 20.6687 -10.5010 0.9998 1.0006 0.9992 0.0001 -0.0000 0.0007
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.1
+ --- dist(#3,#1) = 0.48
+ --- dist(#4,#1) = 0.46
+ --- dist(#5,#1) = 0.475
+ --- dist(#6,#1) = 0.475
+ --- dist(#7,#1) = 0.443
+ --- dist(#8,#1) = 0.476
+ --- dist(#9,#1) = 0.486
+ --- dist(#10,#1) = 0.373
+ --- dist(#11,#1) = 0.48
+ --- dist(#12,#1) = 0.43
+ --- dist(#13,#1) = 0.438
+ --- dist(#14,#1) = 0.39
+ --- dist(#15,#1) = 0.476
+ --- dist(#16,#1) = 0.412
+ --- dist(#17,#1) = 0.431
+ --- dist(#18,#1) = 0.446
+ --- dist(#19,#1) = 0.356
+ --- dist(#20,#1) = 0.5
+ --- dist(#21,#1) = 0.43
+ --- dist(#22,#1) = 0.519
+ --- dist(#23,#1) = 0.49
+ --- dist(#24,#1) = 0.502
+ --- dist(#25,#1) = 0.436
+ --- dist(#26,#1) = 0.468
+ --- dist(#27,#1) = 0.477
+ --- dist(#28,#1) = 0.498
+ --- dist(#29,#1) = 0.486
+ --- dist(#30,#1) = 0.423
++ Start refinement #3 on 30 coarse parameter sets
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.88
+ - Smoothing source; radius=1.88
+ !source mask fill: ubot=64 usiz=159.5
+ - retaining old weight image
+ - using 38906 points from base image [use_all=2]
+ * Exit alignment setup routine
+ 32677 total points stored in 67 'TOHD(17.1973)' bloks (0 duplicates)
*[#26935=-0.23117] *[#26944=-0.232084] *[#26959=-0.2323] *[#26962=-0.232318] *[#26964=-0.232417] *[#26965=-0.232543] *[#26966=-0.232579] *[#26967=-0.232897] *[#26968=-0.233239] *[#26970=-0.233374] *[#26971=-0.233673] *[#26972=-0.233862] *[#26973=-0.234107] *[#26976=-0.234183] *[#26979=-0.234351] *[#26982=-0.234738] *[#26989=-0.234754] *[#26991=-0.234778] *[#26993=-0.234835] *[#26994=-0.234875] *[#26995=-0.23489] *[#26998=-0.234922] *[#27003=-0.234952] *[#27008=-0.23499] *[#27022=-0.235079] *[#27024=-0.235114] *[#27025=-0.235125] *[#27026=-0.235141] *[#27029=-0.235189] *[#27030=-0.235209] *[#27033=-0.235232] *[#27034=-0.235249] *[#27037=-0.235325] *[#27038=-0.235358] *[#27039=-0.235442] *[#27044=-0.235475] *[#27045=-0.235495] *[#27046=-0.235521] *[#27047=-0.235525] *[#27049=-0.235599] *[#27050=-0.235614] *[#27052=-0.235647] *[#27054=-0.235672]
+ - param set #1 has cost=-0.235672 [o=-1 t=-1]
+ -- Parameters = 86.4793 -3.3380 0.0217 -0.7549 -2.1405 -0.4338 0.9891 1.0267 0.9753 -0.0112 0.0131 -0.0443
+ - param set #2 has cost=-0.234666 [o=0 t=0]
+ -- Parameters = 86.4372 -3.2322 -0.0269 -0.6424 -1.6709 -0.2772 0.9885 1.0248 0.9691 -0.0071 0.0076 -0.0358
+ - param set #3 has cost=-0.025822 [o=42 t=21]
+ -- Parameters = 124.8635 -21.9185 -26.3384 8.9122 -2.8957 11.2486 0.9066 0.9500 0.9146 -0.0262 -0.0428 -0.0111
+ - param set #4 has cost=-0.024015 [o=2 t=2]
+ -- Parameters = 96.9853 -22.6084 36.5331 -10.5646 -42.3502 -25.8919 0.9936 0.9938 0.9909 -0.0015 0.0015 0.0018
+ - param set #5 has cost=-0.020677 [o=19 t=1]
+ -- Parameters = 120.2836 31.7782 37.8573 31.4903 -11.7839 -1.0432 1.0280 0.9925 0.9857 -0.0018 0.0016 0.0008
+ - param set #6 has cost=-0.021109 [o=11 t=4]
+ -- Parameters = 122.3854 3.2395 37.8890 32.1032 -27.2846 -8.9726 0.9996 1.0025 0.9997 -0.0002 -0.0004 0.0002
+ - param set #7 has cost=-0.021742 [o=81 t=18]
+ -- Parameters = 100.8685 15.8940 -35.1960 -31.0087 7.4956 -4.8791 0.9890 1.0230 0.9984 -0.0014 0.0064 -0.0026
+ - param set #8 has cost=-0.019976 [o=21 t=5]
+ -- Parameters = 115.3360 31.3654 37.9305 28.3074 -10.2193 2.1137 0.9882 0.9720 1.0138 0.0030 0.0001 -0.0012
+ - param set #9 has cost=-0.019055 [o=6 t=7]
+ -- Parameters = 124.6007 -28.5691 20.2957 5.1816 -41.5171 -44.5870 1.0131 0.9665 0.9992 -0.0041 -0.0017 -0.0082
+ - param set #10 has cost=-0.019574 [o=20 t=11]
+ -- Parameters = 99.3866 -29.8615 26.9020 -5.3338 -28.4230 -34.3867 1.0113 0.9855 1.0194 -0.0030 -0.0004 0.0010
+ - param set #11 has cost=-0.019180 [o=59 t=15]
+ -- Parameters = 120.9740 -8.5507 38.2855 35.6054 -28.5926 -2.1032 0.9445 0.9658 0.9799 0.0065 -0.0097 0.0127
+ - param set #12 has cost=-0.018566 [o=61 t=20]
+ -- Parameters = 95.1214 -16.0331 34.2759 18.9140 -38.5791 -27.1663 0.9916 1.0140 0.9960 0.0012 0.0011 -0.0035
+ - param set #13 has cost=-0.018186 [o=13 t=14]
+ -- Parameters = 94.6948 -15.8655 35.0391 15.9754 -38.3484 -28.1206 0.9895 1.0072 0.9927 -0.0019 0.0009 -0.0027
+ - param set #14 has cost=-0.018618 [o=12 t=24]
+ -- Parameters = 90.7229 19.1250 31.4046 30.8607 -12.3845 -26.7735 0.9996 0.9886 0.9229 -0.0090 0.0068 -0.0166
+ - param set #15 has cost=-0.018967 [o=27 t=3]
+ -- Parameters = 124.5882 -14.9891 37.9295 33.0540 -31.2298 -4.2381 1.0065 1.0007 0.9931 -0.0009 0.0053 0.0016
+ - param set #16 has cost=-0.017468 [o=8 t=25]
+ -- Parameters = 92.1824 -0.8246 28.2401 -37.9242 16.3884 -33.9570 0.9830 0.9798 0.9728 -0.0014 -0.0014 -0.0006
+ - param set #17 has cost=-0.018075 [o=70 t=17]
+ -- Parameters = 86.9585 -14.1999 -34.6419 -5.8658 10.1152 3.4199 0.9923 0.9807 0.9588 0.0087 0.0037 -0.0025
+ - param set #18 has cost=-0.016481 [o=87 t=8]
+ -- Parameters = 80.8196 10.8792 31.9415 39.1021 -23.3509 -12.5253 0.9963 0.9912 0.9875 -0.0009 -0.0018 0.0008
+ - param set #19 has cost=-0.017711 [o=3 t=27]
+ -- Parameters = 92.2159 -30.5634 24.8590 5.7157 -33.5922 -32.8620 0.9876 0.9841 0.9895 0.0194 0.0052 -0.0038
+ - param set #20 has cost=-0.021740 [o=76 t=6]
+ -- Parameters = 83.8861 4.6151 -39.5617 21.7046 10.7784 4.3335 1.0048 1.0034 1.0216 -0.0014 -0.0017 0.0062
+ - param set #21 has cost=-0.015678 [o=23 t=9]
+ -- Parameters = 88.9620 -11.8477 -34.5045 -12.1120 11.6847 4.7198 0.9995 0.9920 0.9895 0.0145 0.0006 -0.0002
+ - param set #22 has cost=-0.016589 [o=68 t=10]
+ -- Parameters = 94.9995 11.4811 -41.7072 -27.1686 18.0982 -14.7609 0.9953 1.0045 1.0033 0.0090 -0.0015 -0.0005
+ - param set #23 has cost=-0.016828 [o=33 t=16]
+ -- Parameters = 110.5268 17.5289 -34.6249 -44.8163 11.5456 22.9792 0.9797 1.0014 0.9661 -0.0008 0.0213 0.0032
+ - param set #24 has cost=-0.016103 [o=55 t=13]
+ -- Parameters = 101.9900 23.5558 -40.5738 -5.7638 -15.8114 40.9105 1.0031 0.9992 1.0094 0.0007 -0.0004 -0.0003
+ - param set #25 has cost=-0.014262 [o=26 t=12]
+ -- Parameters = 112.7557 -14.0757 -34.9144 -28.0999 24.1661 4.0230 0.9951 1.0018 1.0066 0.0048 0.0082 -0.0003
+ - param set #26 has cost=-0.012861 [o=24 t=23]
+ -- Parameters = 119.4894 -7.0076 -37.7482 -11.0534 5.0314 -12.8294 0.9963 1.0036 0.9832 -0.0072 0.0027 -0.0031
+ - param set #27 has cost=-0.017379 [o=43 t=26]
+ -- Parameters = 66.9122 8.7775 -38.5261 20.0114 18.3468 -6.1556 0.9969 0.9943 0.9711 0.0009 0.0066 0.0213
+ - param set #28 has cost=-0.012238 [o=15 t=22]
+ -- Parameters = 119.3286 0.1633 -40.0271 -13.9141 11.3075 -14.2199 0.9960 0.9968 1.0031 0.0023 -0.0007 -0.0006
+ - param set #29 has cost=-0.013300 [o=53 t=28]
+ -- Parameters = 118.9961 -8.2432 -38.8829 -34.5285 24.2673 -0.6308 1.0005 1.0040 1.0051 -0.0013 0.0007 0.0002
+ - param set #30 has cost=-0.012264 [o=74 t=19]
+ -- Parameters = 66.4348 14.9373 -34.0290 33.9352 20.6722 -10.4953 0.9986 1.0022 0.9993 0.0001 0.0001 0.0009
+ - sorting parameter sets by cost
+ - scanning for distances from #1
+ --- dist(#2,#1) = 0.0383
+ --- dist(#3,#1) = 0.48
+ --- dist(#4,#1) = 0.456
+ --- dist(#5,#1) = 0.44
+ --- dist(#6,#1) = 0.495
+ --- dist(#7,#1) = 0.473
+ --- dist(#8,#1) = 0.473
+ --- dist(#9,#1) = 0.474
+ --- dist(#10,#1) = 0.377
+ --- dist(#11,#1) = 0.478
+ --- dist(#12,#1) = 0.491
+ --- dist(#13,#1) = 0.476
+ --- dist(#14,#1) = 0.392
+ --- dist(#15,#1) = 0.428
+ --- dist(#16,#1) = 0.438
+ --- dist(#17,#1) = 0.433
+ --- dist(#18,#1) = 0.36
+ --- dist(#19,#1) = 0.413
+ --- dist(#20,#1) = 0.482
+ --- dist(#21,#1) = 0.49
+ --- dist(#22,#1) = 0.522
+ --- dist(#23,#1) = 0.443
+ --- dist(#24,#1) = 0.507
+ --- dist(#25,#1) = 0.432
+ --- dist(#26,#1) = 0.437
+ --- dist(#27,#1) = 0.486
+ --- dist(#28,#1) = 0.472
+ --- dist(#29,#1) = 0.426
+ --- dist(#30,#1) = 0.501
+ - Total coarse refinement net CPU time = 0.0 s; 9153 funcs
++ *** Fine pass begins ***
+ * Enter alignment setup routine
+ - Smoothing base; radius=1.00
+ - Smoothing source; radius=1.00
+ !source mask fill: ubot=64 usiz=159.5
+ - retaining old weight image
+ * Exit alignment setup routine
++ Picking best parameter set out of 31 cases
+ 32726 total points stored in 68 'TOHD(17.1233)' bloks (0 duplicates)
+ - cost(#1)=-0.229302 * [o=-1 t=-1]
+ -- Parameters = 86.4793 -3.3380 0.0217 -0.7549 -2.1405 -0.4338 0.9891 1.0267 0.9753 -0.0112 0.0131 -0.0443
+ - cost(#2)=-0.228654 [o=0 t=0]
+ -- Parameters = 86.4372 -3.2322 -0.0269 -0.6424 -1.6709 -0.2772 0.9885 1.0248 0.9691 -0.0071 0.0076 -0.0358
+ - cost(#3)=-0.020796 [o=42 t=21]
+ -- Parameters = 124.8635 -21.9185 -26.3384 8.9122 -2.8957 11.2486 0.9066 0.9500 0.9146 -0.0262 -0.0428 -0.0111
+ - cost(#4)=-0.021017 [o=2 t=2]
+ -- Parameters = 96.9853 -22.6084 36.5331 -10.5646 -42.3502 -25.8919 0.9936 0.9938 0.9909 -0.0015 0.0015 0.0018
+ - cost(#5)=-0.019426 [o=81 t=18]
+ -- Parameters = 100.8685 15.8940 -35.1960 -31.0087 7.4956 -4.8791 0.9890 1.0230 0.9984 -0.0014 0.0064 -0.0026
+ - cost(#6)=-0.019844 [o=76 t=6]
+ -- Parameters = 83.8861 4.6151 -39.5617 21.7046 10.7784 4.3335 1.0048 1.0034 1.0216 -0.0014 -0.0017 0.0062
+ - cost(#7)=-0.018617 [o=11 t=4]
+ -- Parameters = 122.3854 3.2395 37.8890 32.1032 -27.2846 -8.9726 0.9996 1.0025 0.9997 -0.0002 -0.0004 0.0002
+ - cost(#8)=-0.016587 [o=19 t=1]
+ -- Parameters = 120.2836 31.7782 37.8573 31.4903 -11.7839 -1.0432 1.0280 0.9925 0.9857 -0.0018 0.0016 0.0008
+ - cost(#9)=-0.017151 [o=21 t=5]
+ -- Parameters = 115.3360 31.3654 37.9305 28.3074 -10.2193 2.1137 0.9882 0.9720 1.0138 0.0030 0.0001 -0.0012
+ - cost(#10)=-0.018519 [o=20 t=11]
+ -- Parameters = 99.3866 -29.8615 26.9020 -5.3338 -28.4230 -34.3867 1.0113 0.9855 1.0194 -0.0030 -0.0004 0.0010
+ - cost(#11)=-0.016436 [o=59 t=15]
+ -- Parameters = 120.9740 -8.5507 38.2855 35.6054 -28.5926 -2.1032 0.9445 0.9658 0.9799 0.0065 -0.0097 0.0127
+ - cost(#12)=-0.016901 [o=6 t=7]
+ -- Parameters = 124.6007 -28.5691 20.2957 5.1816 -41.5171 -44.5870 1.0131 0.9665 0.9992 -0.0041 -0.0017 -0.0082
+ - cost(#13)=-0.016877 [o=27 t=3]
+ -- Parameters = 124.5882 -14.9891 37.9295 33.0540 -31.2298 -4.2381 1.0065 1.0007 0.9931 -0.0009 0.0053 0.0016
+ - cost(#14)=-0.016763 [o=12 t=24]
+ -- Parameters = 90.7229 19.1250 31.4046 30.8607 -12.3845 -26.7735 0.9996 0.9886 0.9229 -0.0090 0.0068 -0.0166
+ - cost(#15)=-0.016364 [o=61 t=20]
+ -- Parameters = 95.1214 -16.0331 34.2759 18.9140 -38.5791 -27.1663 0.9916 1.0140 0.9960 0.0012 0.0011 -0.0035
+ - cost(#16)=-0.015450 [o=13 t=14]
+ -- Parameters = 94.6948 -15.8655 35.0391 15.9754 -38.3484 -28.1206 0.9895 1.0072 0.9927 -0.0019 0.0009 -0.0027
+ - cost(#17)=-0.016063 [o=70 t=17]
+ -- Parameters = 86.9585 -14.1999 -34.6419 -5.8658 10.1152 3.4199 0.9923 0.9807 0.9588 0.0087 0.0037 -0.0025
+ - cost(#18)=-0.015527 [o=3 t=27]
+ -- Parameters = 92.2159 -30.5634 24.8590 5.7157 -33.5922 -32.8620 0.9876 0.9841 0.9895 0.0194 0.0052 -0.0038
+ - cost(#19)=-0.015525 [o=8 t=25]
+ -- Parameters = 92.1824 -0.8246 28.2401 -37.9242 16.3884 -33.9570 0.9830 0.9798 0.9728 -0.0014 -0.0014 -0.0006
+ - cost(#20)=-0.016496 [o=43 t=26]
+ -- Parameters = 66.9122 8.7775 -38.5261 20.0114 18.3468 -6.1556 0.9969 0.9943 0.9711 0.0009 0.0066 0.0213
+ - cost(#21)=-0.014669 [o=33 t=16]
+ -- Parameters = 110.5268 17.5289 -34.6249 -44.8163 11.5456 22.9792 0.9797 1.0014 0.9661 -0.0008 0.0213 0.0032
+ - cost(#22)=-0.015020 [o=68 t=10]
+ -- Parameters = 94.9995 11.4811 -41.7072 -27.1686 18.0982 -14.7609 0.9953 1.0045 1.0033 0.0090 -0.0015 -0.0005
+ - cost(#23)=-0.014942 [o=87 t=8]
+ -- Parameters = 80.8196 10.8792 31.9415 39.1021 -23.3509 -12.5253 0.9963 0.9912 0.9875 -0.0009 -0.0018 0.0008
+ - cost(#24)=-0.014574 [o=55 t=13]
+ -- Parameters = 101.9900 23.5558 -40.5738 -5.7638 -15.8114 40.9105 1.0031 0.9992 1.0094 0.0007 -0.0004 -0.0003
+ - cost(#25)=-0.014193 [o=23 t=9]
+ -- Parameters = 88.9620 -11.8477 -34.5045 -12.1120 11.6847 4.7198 0.9995 0.9920 0.9895 0.0145 0.0006 -0.0002
+ - cost(#26)=-0.012148 [o=26 t=12]
+ -- Parameters = 112.7557 -14.0757 -34.9144 -28.0999 24.1661 4.0230 0.9951 1.0018 1.0066 0.0048 0.0082 -0.0003
+ - cost(#27)=-0.012000 [o=53 t=28]
+ -- Parameters = 118.9961 -8.2432 -38.8829 -34.5285 24.2673 -0.6308 1.0005 1.0040 1.0051 -0.0013 0.0007 0.0002
+ - cost(#28)=-0.011452 [o=24 t=23]
+ -- Parameters = 119.4894 -7.0076 -37.7482 -11.0534 5.0314 -12.8294 0.9963 1.0036 0.9832 -0.0072 0.0027 -0.0031
+ - cost(#29)=-0.011364 [o=74 t=19]
+ -- Parameters = 66.4348 14.9373 -34.0290 33.9352 20.6722 -10.4953 0.9986 1.0022 0.9993 0.0001 0.0001 0.0009
+ - cost(#30)=-0.010783 [o=15 t=22]
+ -- Parameters = 119.3286 0.1633 -40.0271 -13.9141 11.3075 -14.2199 0.9960 0.9968 1.0031 0.0023 -0.0007 -0.0006
+ - cost(#31)=-0.129342 [o=-2 t=-2]
+ -- Parameters = 86.1901 -3.4407 -1.7088 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
+ -num_rtb 99 ==> refine all 31 cases
+ - cost(#1)=-0.229705 * [o=-1 t=-1]
+ -- Parameters = 86.4786 -3.3794 0.0456 -0.7344 -2.0985 -0.4671 0.9912 1.0266 0.9763 -0.0110 0.0121 -0.0449
+ - cost(#2)=-0.229554 [o=0 t=0]
+ -- Parameters = 86.4108 -3.3549 0.0075 -0.6847 -1.9108 -0.2988 0.9897 1.0254 0.9729 -0.0093 0.0067 -0.0411
+ - cost(#3)=-0.020800 [o=42 t=21]
+ -- Parameters = 124.8687 -21.9275 -26.3385 8.9060 -2.8977 11.2572 0.9066 0.9499 0.9145 -0.0261 -0.0428 -0.0111
+ - cost(#4)=-0.021932 [o=2 t=2]
+ -- Parameters = 96.7132 -21.8547 37.0594 -11.0616 -41.9192 -26.4843 0.9931 0.9931 0.9950 -0.0013 0.0013 0.0069
+ - cost(#5)=-0.019536 [o=81 t=18]
+ -- Parameters = 100.8433 15.8919 -35.1646 -30.9880 7.4855 -4.8544 0.9870 1.0216 0.9993 -0.0015 0.0070 -0.0025
+ - cost(#6)=-0.021827 [o=76 t=6]
+ -- Parameters = 82.3364 4.8772 -39.6003 20.1755 10.4514 3.8632 1.0049 1.0059 1.0260 -0.0085 -0.0025 0.0079
+ - cost(#7)=-0.019391 [o=11 t=4]
+ -- Parameters = 122.1885 3.3850 37.7804 31.9993 -26.7764 -9.1048 1.0047 1.0043 1.0237 -0.0004 -0.0009 0.0010
+ - cost(#8)=-0.016887 [o=19 t=1]
+ -- Parameters = 120.3862 31.8744 37.9044 31.2181 -11.7938 -1.2006 1.0284 0.9884 0.9874 -0.0101 0.0025 0.0006
+ - cost(#9)=-0.017390 [o=21 t=5]
+ -- Parameters = 115.3469 31.2859 37.8570 28.2962 -10.3465 2.2030 0.9873 0.9648 1.0135 0.0016 0.0010 -0.0018
+ - cost(#10)=-0.018693 [o=20 t=11]
+ -- Parameters = 99.3262 -29.9447 26.9371 -5.3159 -28.4886 -34.0726 1.0107 0.9849 1.0213 -0.0036 -0.0003 0.0007
+ - cost(#11)=-0.016689 [o=59 t=15]
+ -- Parameters = 120.8994 -8.5654 38.2884 35.5660 -28.3844 -2.2898 0.9449 0.9652 0.9808 0.0065 -0.0097 0.0202
+ - cost(#12)=-0.017087 [o=6 t=7]
+ -- Parameters = 124.5712 -28.6409 20.3221 5.1349 -41.6487 -44.5124 1.0125 0.9652 0.9978 -0.0043 -0.0015 -0.0151
+ - cost(#13)=-0.018747 [o=27 t=3]
+ -- Parameters = 124.8220 -15.4132 35.7434 32.8513 -32.2128 -4.2913 1.0051 1.0020 0.9925 -0.0014 0.0061 0.0027
+ - cost(#14)=-0.016958 [o=12 t=24]
+ -- Parameters = 90.6424 19.0892 31.4461 30.8611 -12.4391 -26.9602 1.0001 0.9900 0.9213 -0.0096 0.0069 -0.0164
+ - cost(#15)=-0.016621 [o=61 t=20]
+ -- Parameters = 95.2265 -16.0288 34.3051 18.8769 -38.6949 -27.3742 0.9949 1.0116 0.9938 0.0095 0.0005 -0.0030
+ - cost(#16)=-0.015480 [o=13 t=14]
+ -- Parameters = 94.6942 -15.8722 35.0063 16.0018 -38.3548 -28.1013 0.9892 1.0073 0.9938 -0.0018 0.0006 -0.0023
+ - cost(#17)=-0.017580 [o=70 t=17]
+ -- Parameters = 87.0306 -14.2621 -34.6583 -9.0179 9.8610 3.8227 0.9879 0.9809 0.9564 0.0087 0.0030 -0.0024
+ - cost(#18)=-0.016272 [o=3 t=27]
+ -- Parameters = 92.1936 -30.6225 24.8890 5.7653 -33.6379 -32.8354 0.9906 0.9904 0.9919 0.0296 0.0046 -0.0038
+ - cost(#19)=-0.018831 [o=8 t=25]
+ -- Parameters = 92.0665 -2.7610 29.1632 -37.9380 15.4778 -34.5080 0.9731 0.9648 0.9439 -0.0023 -0.0035 0.0016
+ - cost(#20)=-0.016643 [o=43 t=26]
+ -- Parameters = 66.9145 8.7317 -38.5499 19.9377 18.3452 -6.1108 1.0030 0.9976 0.9713 0.0006 0.0051 0.0221
+ - cost(#21)=-0.015076 [o=33 t=16]
+ -- Parameters = 110.5064 17.5324 -34.6949 -44.8090 11.7870 22.7514 0.9787 1.0007 0.9652 -0.0008 0.0293 0.0042
+ - cost(#22)=-0.015064 [o=68 t=10]
+ -- Parameters = 94.9821 11.4850 -41.7087 -27.1684 18.0621 -14.7438 0.9953 1.0047 1.0033 0.0097 -0.0014 -0.0005
+ - cost(#23)=-0.015215 [o=87 t=8]
+ -- Parameters = 80.8903 10.8767 31.8345 38.9750 -23.2805 -12.6024 0.9964 0.9868 0.9912 -0.0075 -0.0002 0.0015
+ - cost(#24)=-0.014807 [o=55 t=13]
+ -- Parameters = 101.9003 23.5725 -40.5280 -5.6755 -15.6652 40.9313 1.0032 0.9991 1.0112 0.0085 -0.0004 -0.0001
+ - cost(#25)=-0.014391 [o=23 t=9]
+ -- Parameters = 88.9471 -11.7913 -34.3018 -12.1401 11.8714 4.7590 1.0001 0.9921 0.9903 0.0145 0.0001 -0.0000
+ - cost(#26)=-0.012283 [o=26 t=12]
+ -- Parameters = 112.7510 -14.0798 -34.9248 -28.0911 24.1450 4.0337 0.9952 1.0025 1.0059 0.0123 0.0083 -0.0004
+ - cost(#27)=-0.012050 [o=53 t=28]
+ -- Parameters = 119.0245 -8.2729 -38.8807 -34.4827 24.2436 -0.6653 1.0003 1.0042 1.0074 -0.0013 0.0015 0.0001
+ - cost(#28)=-0.011891 [o=24 t=23]
+ -- Parameters = 119.1164 -6.9467 -37.4876 -11.0812 5.1131 -12.3826 0.9973 1.0034 0.9823 -0.0065 0.0021 -0.0026
+ - cost(#29)=-0.011366 [o=74 t=19]
+ -- Parameters = 66.4278 14.9361 -34.0231 33.9244 20.6755 -10.4931 0.9986 1.0021 0.9994 0.0001 0.0002 0.0008
+ - cost(#30)=-0.012654 [o=15 t=22]
+ -- Parameters = 118.6806 -0.2779 -39.9209 -10.4236 10.9901 -14.6070 0.9859 0.9982 1.0056 0.0036 -0.0026 0.0003
+ - cost(#31)=-0.226142 [o=-2 t=-2]
+ -- Parameters = 86.2853 -3.2417 0.0041 -0.6043 -1.3783 0.2327 0.9846 1.0247 0.9738 -0.0073 -0.0072 -0.0283
+ - case #1 [o=-1 t=-1] is now the best
+ - Initial cost = -0.229705
+ - Initial fine Parameters = 86.4786 -3.3794 0.0456 -0.7344 -2.0985 -0.4671 0.9912 1.0266 0.9763 -0.0110 0.0121 -0.0449
+ - Finalish cost = -0.230087 ; 443 funcs
+ - ini Finalish Parameters = 86.4243 -3.3934 0.0579 -0.6907 -2.4008 -0.2686 0.9907 1.0261 0.9755 -0.0090 0.0076 -0.0512
+ - Final cost = -0.230087 ; 283 funcs
+ Final fine fit Parameters:
x-shift= 86.4239 y-shift= -3.3912 z-shift= 0.0590 ... enorm= 86.4904 mm
z-angle= -0.6917 x-angle= -2.3988 y-angle= -0.2681 ... total= 2.5094 deg
x-scale= 0.9907 y-scale= 1.0261 z-scale= 0.9755 ... vol3D= 0.9917 = base bigger than source
y/x-shear= -0.0090 z/x-shear= 0.0076 z/y-shear= -0.0512
+ - Fine net CPU time = 0.0 s
++ Computing output image
++ image warp: parameters = 86.4239 -3.3912 0.0590 -0.6917 -2.3988 -0.2681 0.9907 1.0261 0.9755 -0.0090 0.0076 -0.0512
++ Unloading unneeded data
++ Output dataset ./sub-08_T1w_al_junk+orig.BRIK
++ Wrote -1Dmatrix_save ./sub-08_T1w_al_junk_mat.aff12.1D
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 17.3
++ ###########################################################
++ # PLEASE check results VISUALLY for alignment quality #
++ ###########################################################
#++ Creating final output: skullstripped anat data
copying from dataset /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_processing/sub_08.results/__tt_sub-08_T1w_ns+orig to /home/jovyan/Git_repositories/example-notebooks/books/functional_imaging/afni_processing/sub_08.results/sub-08_T1w_ns+orig
#Script is running (command trimmed):
3dcopy ./__tt_sub-08_T1w_ns+orig sub-08_T1w_ns
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#++ Creating final output: anat data aligned to epi
# copy is not necessary
#++ Saving history
#Script is running (command trimmed):
3dNotes -h "align_epi_anat.py -anat2epi -anat sub-08_T1w+orig \
-save_skullstrip -suffix _al_junk -epi vr_base_min_outlier+orig -epi_base \
0 -epi_strip 3dAutomask -giant_move -volreg off -tshift off" \
./sub-08_T1w_al_junk+orig
#++ Removing all the temporary files
#Script is running:
\rm -f ./__tt_vr_base_min_outlier*
#Script is running:
\rm -f ./__tt_sub-08_T1w*
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
# Finished alignment successfully
@auto_tlrc -base MNI_avg152T1+tlrc -input sub-08_T1w_ns+orig -no_ss -init_xform AUTO_CENTER
Performing center alignment with @Align_Centers
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset ./sub-08_T1w_ns_shft+orig
+ deoblique
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename sub-08_T1w_ns_shft+orig.HEAD -> __ats_tmp__sub-08_T1w_ns_shft+orig.HEAD
++ THD_rename_dataset_files: rename sub-08_T1w_ns_shft+orig.BRIK -> __ats_tmp__sub-08_T1w_ns_shft+orig.BRIK
** THD_rename_dataset_files: old header sub-08_T1w_ns_shft+acpc.HEAD doesn't exist!
** THD_rename_dataset_files: old header sub-08_T1w_ns_shft+tlrc.HEAD doesn't exist!
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Center distance of 0.000000 mm
Padding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp__ref_MNI_avg152T1_15pad+tlrc.BRIK
Resampling ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp_step+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__resamp+tlrc.BRIK
Clipping -0.000100 1336.000100 ...
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./__ats_tmp___rs_pre.sub-08_T1w_ns+tlrc.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___rs_pre.sub-08_T1w_ns+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_NN+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp__resamp_edge_art+tlrc.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Registration (cubic final interpolation) ...
++ 3dWarpDrive: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Initial scale factor set to 0.30/125.00=4.2e+02
RMS[0] = 0.283712 0.155217 ITER = 8/50
0.283712
Warping has converged.
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
Applying brain mask
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp__reg_braintlrcstep+orig.BRIK
++ 3dWarp: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-08_T1w_ns+orig.HEAD -> __ats_tmp___pad15_pre.sub-08_T1w_ns.skl+orig.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___pad15_pre.sub-08_T1w_ns+orig.BRIK -> __ats_tmp___pad15_pre.sub-08_T1w_ns.skl+orig.BRIK
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ././__ats_tmp___pad15_pre.sub-08_T1w_ns+orig.BRIK
Unpadding ...
++ 3dZeropad: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ output dataset: ./__ats_tmp___upad15_pre.sub-08_T1w_ns+orig.BRIK
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-08_T1w_ns+orig
*+ WARNING: Changing the space of an ORIG view dataset may cause confusion!
*+ WARNING: NIFTI copies will be interpreted as TLRC view (not TLRC space).
*+ WARNING: Consider changing the view of the dataset to TLRC view also
++ 3drefit processed 1 datasets
Changing view of transformed anatomy
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-08_T1w_ns+orig.HEAD
+ changing dataset view code
+ Changed dataset view type and filenames.
++ 3drefit processed 1 datasets
Setting parent with 3drefit -wset sub-08_T1w_ns+orig __ats_tmp___upad15_pre.sub-08_T1w_ns+tlrc
++ 3drefit: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Processing AFNI dataset __ats_tmp___upad15_pre.sub-08_T1w_ns+tlrc
+ setting Warp parent
++ 3drefit processed 1 datasets
++ 3drename: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Warning: ignoring +tlrc on new_prefix.
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-08_T1w_ns+tlrc.HEAD -> sub-08_T1w_ns+tlrc.HEAD
++ THD_rename_dataset_files: rename __ats_tmp___upad15_pre.sub-08_T1w_ns+tlrc.BRIK -> sub-08_T1w_ns+tlrc.BRIK
Cleanup ...
cat_matvec sub-08_T1w_ns+tlrc::WARP_DATA -I
if ( ! -f sub-08_T1w_ns+tlrc.HEAD ) then
foreach run ( 01 02 )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r01.1D -prefix rm.epi.volreg.r01 -cubic -1Dmatrix_save mat.r01.vr.aff12.1D pb01.sub_08.r01.tshift+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb01.sub_08.r01.tshift+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ Automask has 40918 voxels
+ 5959 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=-0.014 pitch=-0.044 yaw=-0.059 dS=-0.092 dL=-0.041 dP=-0.083
++ Mean: roll=+0.022 pitch=+0.035 yaw=+0.065 dS=+0.016 dL=+0.029 dP=+0.013
++ Max : roll=+0.119 pitch=+0.184 yaw=+0.253 dS=+0.181 dL=+0.123 dP=+0.235
++ Max displacements (mm) for each sub-brick:
0.15(0.00) 0.09(0.16) 0.06(0.09) 0.05(0.08) 0.07(0.08) 0.07(0.03) 0.08(0.05) 0.06(0.05) 0.15(0.13) 0.09(0.12) 0.15(0.12) 0.09(0.13) 0.11(0.10) 0.09(0.06) 0.09(0.05) 0.11(0.04) 0.04(0.10) 0.08(0.08) 0.07(0.05) 0.13(0.10) 0.06(0.11) 0.06(0.09) 0.07(0.09) 0.03(0.05) 0.09(0.07) 0.06(0.13) 0.07(0.09) 0.05(0.08) 0.00(0.05) 0.06(0.06) 0.06(0.08) 0.05(0.06) 0.10(0.07) 0.06(0.06) 0.07(0.09) 0.06(0.10) 0.08(0.08) 0.08(0.07) 0.12(0.10) 0.08(0.11) 0.09(0.08) 0.11(0.07) 0.05(0.08) 0.08(0.05) 0.03(0.08) 0.07(0.06) 0.08(0.09) 0.06(0.05) 0.10(0.07) 0.08(0.09) 0.12(0.10) 0.10(0.08) 0.06(0.07) 0.14(0.14) 0.09(0.11) 0.11(0.10) 0.14(0.06) 0.09(0.12) 0.12(0.11) 0.10(0.09) 0.10(0.06) 0.15(0.07) 0.09(0.12) 0.11(0.10) 0.09(0.15) 0.14(0.16) 0.09(0.09) 0.10(0.03) 0.11(0.07) 0.08(0.07) 0.13(0.11) 0.09(0.11) 0.13(0.11) 0.09(0.08) 0.10(0.06) 0.13(0.10) 0.10(0.09) 0.14(0.11) 0.11(0.14) 0.11(0.10) 0.09(0.07) 0.10(0.08) 0.11(0.06) 0.10(0.10) 0.16(0.16) 0.10(0.16) 0.13(0.14) 0.13(0.11) 0.17(0.07) 0.18(0.07) 0.15(0.09) 0.15(0.07) 0.12(0.10) 0.17(0.10) 0.15(0.05) 0.16(0.07) 0.14(0.06) 0.16(0.10) 0.24(0.15) 0.14(0.17) 0.21(0.14) 0.16(0.12) 0.29(0.22) 0.20(0.16) 0.23(0.11) 0.21(0.07) 0.21(0.10) 0.23(0.05) 0.19(0.12) 0.22(0.08) 0.18(0.09) 0.23(0.10) 0.19(0.11) 0.28(0.14) 0.21(0.10) 0.23(0.06) 0.23(0.07) 0.29(0.15) 0.22(0.17) 0.29(0.10) 0.30(0.04) 0.27(0.07) 0.31(0.08) 0.28(0.06) 0.34(0.11) 0.30(0.07) 0.37(0.11) 0.28(0.15) 0.36(0.14) 0.29(0.14) 0.36(0.15) 0.32(0.30) 0.41(0.27) 0.49(0.29) 0.46(0.11) 0.49(0.18) 0.56(0.18) 0.55(0.14) 0.55(0.10) 0.54(0.03) 0.52(0.12) 0.51(0.14) 0.48(0.15) 0.41(0.17) 0.41(0.22) 0.42(0.12)
++ Max displacement in automask = 0.56 (mm) at sub-brick 136
++ Max delta displ in automask = 0.30 (mm) at sub-brick 131
++ Wrote dataset to disk in ./rm.epi.volreg.r01+orig.BRIK
3dcalc -overwrite -a pb01.sub_08.r01.tshift+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-08_T1w_ns+tlrc::WARP_DATA -I sub-08_T1w_al_junk_mat.aff12.1D -I mat.r01.vr.aff12.1D
3dAllineate -base sub-08_T1w_ns+tlrc -input pb01.sub_08.r01.tshift+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb01.sub_08.r01.tshift+orig.HEAD
++ Base dataset: ./sub-08_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 0.027 -8.241 -29.822
+ shift search range is +/- = 57.780 69.336 57.780
+ 0.0% 11.9% 51.6%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.4
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-08_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r01.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r01
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 1.280 -18.654 -32.063
+ shift search range is +/- = 57.780 69.336 57.780
+ 2.2% 26.9% 55.5%
++ Output dataset ./rm.epi.1.r01+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 3.8
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r01 rm.epi.1.r01+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.epi.min.r01+tlrc.BRIK
end
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig -1Dfile dfile.r02.1D -prefix rm.epi.volreg.r02 -cubic -1Dmatrix_save mat.r02.vr.aff12.1D pb01.sub_08.r02.tshift+orig
++ 3dvolreg: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: RW Cox
++ Reading in base dataset ./vr_base_min_outlier+orig.BRIK
++ Reading input dataset ./pb01.sub_08.r02.tshift+orig.BRIK
++ Edging: x=3 y=3 z=2
++ Creating mask for -maxdisp
+ Automask has 40888 voxels
+ 5952 voxels left in -maxdisp mask after erosion
++ Initializing alignment base
++ Starting final pass on 146 sub-bricks: 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145..
++ CPU time for realignment=0 s [=0 s/sub-brick]
++ Min : roll=+0.065 pitch=-0.196 yaw=-0.295 dS=-0.284 dL=+0.045 dP=-0.220
++ Mean: roll=+0.160 pitch=+0.114 yaw=+0.164 dS=-0.032 dL=+0.132 dP=-0.070
++ Max : roll=+0.233 pitch=+0.250 yaw=+0.335 dS=+0.142 dL=+0.306 dP=+0.067
++ Max displacements (mm) for each sub-brick:
0.70(0.00) 0.61(0.16) 0.62(0.09) 0.60(0.07) 0.58(0.06) 0.63(0.08) 0.61(0.07) 0.62(0.08) 0.60(0.07) 0.55(0.07) 0.62(0.08) 0.61(0.11) 0.61(0.07) 0.59(0.05) 0.50(0.10) 0.54(0.06) 0.52(0.08) 0.57(0.09) 0.58(0.09) 0.56(0.09) 0.58(0.06) 0.56(0.07) 0.59(0.10) 0.57(0.09) 0.57(0.07) 0.59(0.05) 0.56(0.05) 0.59(0.08) 0.60(0.08) 0.58(0.07) 0.58(0.07) 0.58(0.06) 0.58(0.02) 0.59(0.05) 0.57(0.34) 0.38(0.39) 0.47(0.48) 0.90(0.47) 0.59(0.35) 0.49(0.19) 0.50(0.16) 0.46(0.12) 0.48(0.16) 0.46(0.17) 0.48(0.13) 0.47(0.10) 0.48(0.10) 0.49(0.09) 0.51(0.10) 0.53(0.06) 0.51(0.09) 0.53(0.09) 0.50(0.10) 0.50(0.09) 0.50(0.08) 0.49(0.07) 0.52(0.06) 0.46(0.10) 0.46(0.21) 0.52(0.33) 0.50(0.20) 0.54(0.12) 0.54(0.09) 0.60(0.12) 0.55(0.14) 0.59(0.12) 0.58(0.10) 0.62(0.13) 0.53(0.11) 0.49(0.14) 0.49(0.04) 0.46(0.07) 0.55(0.11) 0.48(0.14) 0.57(0.15) 0.46(0.14) 0.54(0.11) 0.44(0.13) 0.51(0.11) 0.47(0.10) 0.55(0.15) 0.59(0.11) 0.56(0.13) 0.53(0.13) 0.60(0.14) 0.58(0.10) 0.55(0.04) 0.53(0.07) 0.59(0.11) 0.49(0.15) 0.57(0.14) 0.52(0.17) 0.77(0.46) 0.58(0.27) 0.57(0.20) 0.57(0.10) 0.58(0.13) 0.56(0.14) 0.57(0.14) 0.55(0.09) 0.60(0.10) 0.55(0.09) 0.55(0.05) 0.55(0.04) 0.57(0.07) 0.61(0.06) 0.53(0.11) 0.67(0.16) 0.55(0.16) 0.59(0.11) 0.53(0.10) 0.54(0.06) 0.57(0.07) 0.51(0.12) 0.56(0.12) 0.57(0.17) 0.64(0.20) 0.58(0.21) 0.56(0.09) 0.60(0.07) 0.55(0.07) 0.61(0.08) 0.56(0.12) 0.60(0.14) 0.59(0.10) 0.61(0.07) 0.59(0.06) 0.56(0.08) 0.67(0.14) 0.61(0.12) 0.64(0.14) 0.64(0.04) 0.64(0.19) 0.57(0.25) 0.54(0.10) 0.54(0.07) 0.53(0.05) 0.57(0.11) 0.53(0.11) 0.58(0.12) 0.53(0.09) 0.55(0.06) 0.52(0.11) 0.56(0.13) 0.55(0.10) 0.53(0.06)
++ Max displacement in automask = 0.90 (mm) at sub-brick 37
++ Max delta displ in automask = 0.48 (mm) at sub-brick 36
++ Wrote dataset to disk in ./rm.epi.volreg.r02+orig.BRIK
3dcalc -overwrite -a pb01.sub_08.r02.tshift+orig -expr 1 -prefix rm.epi.all1
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
*+ WARNING: input 'a' is not used in the expression
++ Output dataset ./rm.epi.all1+orig.BRIK
cat_matvec -ONELINE sub-08_T1w_ns+tlrc::WARP_DATA -I sub-08_T1w_al_junk_mat.aff12.1D -I mat.r02.vr.aff12.1D
3dAllineate -base sub-08_T1w_ns+tlrc -input pb01.sub_08.r02.tshift+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -prefix rm.epi.nomask.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./pb01.sub_08.r02.tshift+orig.HEAD
++ Base dataset: ./sub-08_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = -0.046 -8.175 -29.810
+ shift search range is +/- = 57.780 69.336 57.780
+ 0.1% 11.8% 51.6%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 146 sub-bricks ==========
volume 0..1..2..3..4..5..6..7..8..9..10..11..12..13..14..15..16..17..18..19..20..21..22..23..24..25..26..27..28..29..30..31..32..33..34..35..36..37..38..39..40..41..42..43..44..45..46..47..48..49..50..51..52..53..54..55..56..57..58..59..60..61..62..63..64..65..66..67..68..69..70..71..72..73..74..75..76..77..78..79..80..81..82..83..84..85..86..87..88..89..90..91..92..93..94..95..96..97..98..99..100..101..102..103..104..105..106..107..108..109..110..111..112..113..114..115..116..117..118..119..120..121..122..123..124..125..126..127..128..129..130..131..132..133..134..135..136..137..138..139..140..141..142..143..144..145
++ Output dataset ./rm.epi.nomask.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 4.5
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dAllineate -base sub-08_T1w_ns+tlrc -input rm.epi.all1+orig -1Dmatrix_apply mat.r02.warp.aff12.1D -mast_dxyz 3 -final NN -quiet -prefix rm.epi.1.r02
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 1.280 -18.654 -32.063
+ shift search range is +/- = 57.780 69.336 57.780
+ 2.2% 26.9% 55.5%
++ Output dataset ./rm.epi.1.r02+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 3.9
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dTstat -min -prefix rm.epi.min.r02 rm.epi.1.r02+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.epi.min.r02+tlrc.BRIK
end
cat dfile.r01.1D dfile.r02.1D
1d_tool.py -infile dfile_rall.1D -set_nruns 2 -derivative -collapse_cols euclidean_norm -write motion_sub_08_enorm.1D
/opt/afni-latest/afnipy/lib_afni1D.py:1302: SyntaxWarning: 'str' object is not callable; perhaps you missed a comma?
print('** uncensor from vec: nt = %d, but nocen len = %d' \
3dMean -datum short -prefix rm.epi.mean rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.HEAD
++ 3dMean: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dcalc -a rm.epi.mean+tlrc -expr step(a-0.999) -prefix mask_epi_extents
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./mask_epi_extents+tlrc.BRIK
foreach run ( 01 02 )
3dcalc -a rm.epi.nomask.r01+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb02.sub_08.r01.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb02.sub_08.r01.volreg+tlrc.BRIK
end
3dcalc -a rm.epi.nomask.r02+tlrc -b mask_epi_extents+tlrc -expr a*b -prefix pb02.sub_08.r02.volreg
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb02.sub_08.r02.volreg+tlrc.BRIK
end
cat_matvec -ONELINE sub-08_T1w_ns+tlrc::WARP_DATA -I sub-08_T1w_al_junk_mat.aff12.1D -I
3dAllineate -base sub-08_T1w_ns+tlrc -input vr_base_min_outlier+orig -1Dmatrix_apply mat.basewarp.aff12.1D -mast_dxyz 3 -prefix final_epi_vr_base_min_outlier
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./vr_base_min_outlier+orig.HEAD
++ Base dataset: ./sub-08_T1w_ns+tlrc.HEAD
++ Loading datasets into memory
*+ WARNING: center of mass shifts (-cmass) are turned off, but would be TERRIBLY large!
*+ WARNING: - at least one is more than 50% of search range
+ -cmass x y z shifts = 0.073 -8.252 -29.781
+ shift search range is +/- = 57.780 69.336 57.780
+ 0.1% 11.9% 51.5%
++ master dataset for output = base
++ changing output grid spacing to 3.0000 mm
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ Output dataset ./final_epi_vr_base_min_outlier+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.1
++ ###########################################################
+
++ ***********************************************************
*+ WARNING: -cmass was turned off, but might have been needed :(
+ Please check your results - PLEASE PLEASE PLEASE
++ ***********************************************************
3dcopy sub-08_T1w_ns+tlrc anat_final.sub_08
++ 3dcopy: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
3dAllineate -base final_epi_vr_base_min_outlier+tlrc -allcostX -input anat_final.sub_08+tlrc
tee out.allcostX.txt
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ Source dataset: ./anat_final.sub_08+tlrc.HEAD
++ Base dataset: ./final_epi_vr_base_min_outlier+tlrc.HEAD
++ Loading datasets into memory
++ Local correlation: blok type = 'TOHD(15.531)'
+ -cmass x y z shifts = -0.040 1.128 0.173
+ shift search range is +/- = 66.447 79.929 68.373
*+ WARNING: No output dataset will be calculated
++ OpenMP thread count = 15
+ initial Parameters = 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 1.0000 1.0000 1.0000 0.0000 0.0000 0.0000
++ allcost output: init #0
ls = 0.102416
sp = 0.294745
mi = 2.69858
crM = 0.0196901
nmi = 0.820873
je = 2.69858
hel = -0.12172
crA = 0.141537
crU = 0.160044
lss = 0.897584
lpc = 0.647903
lpa = 0.352097
lpc+ = 0.746398
lpa+ = 0.450592
++ Output dataset ./volumized+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 0.7
++ ###########################################################
3dAllineate -source sub-08_T1w+orig -master anat_final.sub_08+tlrc -final wsinc5 -1Dmatrix_apply warp.anat.Xat.1D -prefix anat_w_skull_warped
++ 3dAllineate: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Zhark the Registrator
++ -1Dmatrix_apply: converting input 3x4 array to 1 row of 12 numbers
++ Source dataset: ./sub-08_T1w+orig.HEAD
++ Base dataset: (not given)
++ Loading datasets into memory
+ -cmass x y z shifts = 0.000 0.000 0.000
+ shift search range is +/- = 56.175 81.855 81.855
++ OpenMP thread count = 15
++ ========== Applying transformation to 1 sub-bricks ==========
++ wsinc5 interpolation setup:
+ taper function = Min sidelobe 3 term
+ taper cut point = 0.000
+ window radius = 5 voxels
+ window shape = Cubical
+ The above can be altered via the AFNI_WSINC5_* environment variables.
+ (To avoid this message, 'setenv AFNI_WSINC5_SILENT YES'.)
+ wsinc5 CUBE(5) mask has 1000 points
++ Output dataset ./anat_w_skull_warped+tlrc.BRIK
++ 3dAllineate: total CPU time = 0.0 sec Elapsed = 1.0
++ ###########################################################
foreach run ( 01 02 )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb03.sub_08.r01.blur pb02.sub_08.r01.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb03.sub_08.r01.blur+tlrc.BRIK
end
3dmerge -1blur_fwhm 4.0 -doall -prefix pb03.sub_08.r02.blur pb02.sub_08.r02.volreg+tlrc
++ 3dmerge: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ default -1dindex = 0
++ default -1tindex = 1
Program 3dmerge
3dmerge: edit and combine 3D datasets, by RW Cox
++ editing input dataset in memory (75.6 MB)
..................................................................................................................................................
-- Wrote edited dataset: ./pb03.sub_08.r02.blur+tlrc.BRIK
end
foreach run ( 01 02 )
3dAutomask -prefix rm.mask_r01 pb03.sub_08.r01.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb03.sub_08.r01.blur+tlrc
++ Forming automask
+ Fixed clip level = 343.540375
+ Used gradual clip level = 322.806030 .. 362.186707
+ Number voxels above clip level = 69428
+ Clustering voxels ...
+ Largest cluster has 68830 voxels
+ Clustering voxels ...
+ Largest cluster has 68733 voxels
+ Filled 139 voxels in small holes; now have 68872 voxels
+ Filled 2 voxels in large holes; now have 68874 voxels
+ Clustering voxels ...
+ Largest cluster has 68873 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 202760 voxels
+ Mask now has 68873 voxels
++ 68873 voxels in the mask [out of 271633: 25.36%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 5 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 8 z-planes are zero [from S]
++ Output dataset ./rm.mask_r01+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dAutomask -prefix rm.mask_r02 pb03.sub_08.r02.blur+tlrc
++ 3dAutomask: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: Emperor Zhark
++ Loading dataset pb03.sub_08.r02.blur+tlrc
++ Forming automask
+ Fixed clip level = 342.217621
+ Used gradual clip level = 321.662537 .. 360.685547
+ Number voxels above clip level = 69449
+ Clustering voxels ...
+ Largest cluster has 68853 voxels
+ Clustering voxels ...
+ Largest cluster has 68749 voxels
+ Filled 134 voxels in small holes; now have 68883 voxels
+ Filled 2 voxels in large holes; now have 68885 voxels
+ Clustering voxels ...
+ Largest cluster has 68885 voxels
+ Clustering non-brain voxels ...
+ Clustering voxels ...
+ Largest cluster has 202748 voxels
+ Mask now has 68885 voxels
++ 68885 voxels in the mask [out of 271633: 25.36%]
++ first 7 x-planes are zero [from L]
++ last 6 x-planes are zero [from R]
++ first 5 y-planes are zero [from P]
++ last 4 y-planes are zero [from A]
++ first 0 z-planes are zero [from I]
++ last 8 z-planes are zero [from S]
++ Output dataset ./rm.mask_r02+tlrc.BRIK
++ CPU time = 0.000000 sec
end
3dmask_tool -inputs rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.HEAD -union -prefix full_mask.sub_08
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 0 over 2 volumes gives min count 0
++ voxel limits: 0 clipped, 68948 survived, 202685 were zero
++ writing result full_mask.sub_08...
++ Output dataset ./full_mask.sub_08+tlrc.BRIK
3dresample -master full_mask.sub_08+tlrc -input sub-08_T1w_ns+tlrc -prefix rm.resam.anat
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.anat+tlrc -prefix mask_anat.sub_08
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 76711 survived, 194922 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_anat.sub_08...
++ Output dataset ./mask_anat.sub_08+tlrc.BRIK
3dmask_tool -input full_mask.sub_08+tlrc mask_anat.sub_08+tlrc -inter -prefix mask_epi_anat.sub_08
++ processing 2 input dataset(s), NN=2...
++ padding all datasets by 0 (for dilations)
++ frac 1 over 2 volumes gives min count 2
++ voxel limits: 10363 clipped, 67648 survived, 193622 were zero
++ writing result mask_epi_anat.sub_08...
++ Output dataset ./mask_epi_anat.sub_08+tlrc.BRIK
3dABoverlap -no_automask full_mask.sub_08+tlrc mask_anat.sub_08+tlrc
tee out.mask_ae_overlap.txt
++ 3dABoverlap: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#A=./full_mask.sub_08+tlrc.BRIK B=./mask_anat.sub_08+tlrc.BRIK
#A #B #(A uni B) #(A int B) #(A \ B) #(B \ A) %(A \ B) %(B \ A) Rx(B/A) Ry(B/A) Rz(B/A)
68948 76711 78011 67648 1300 9063 1.8855 11.8145 1.0652 0.9478 1.1189
3ddot -dodice full_mask.sub_08+tlrc mask_anat.sub_08+tlrc
tee out.mask_ae_dice.txt
0.928854
3dresample -master full_mask.sub_08+tlrc -prefix ./rm.resam.group -input /opt/afni-latest/MNI_avg152T1+tlrc
3dmask_tool -dilate_input 5 -5 -fill_holes -input rm.resam.group+tlrc -prefix mask_group
++ no -frac option: defaulting to -union
++ processing 1 input dataset(s), NN=2...
++ padding all datasets by 5 (for dilations)
++ frac 0 over 1 volumes gives min count 0
++ voxel limits: 0 clipped, 73409 survived, 198224 were zero
++ filled 0 holes (0 voxels)
++ writing result mask_group...
++ Output dataset ./mask_group+tlrc.BRIK
3ddot -dodice mask_anat.sub_08+tlrc mask_group+tlrc
tee out.mask_at_dice.txt
0.960445
foreach run ( 01 02 )
3dTstat -prefix rm.mean_r01 pb03.sub_08.r01.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.mean_r01+tlrc.BRIK
3dcalc -a pb03.sub_08.r01.blur+tlrc -b rm.mean_r01+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb04.sub_08.r01.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb04.sub_08.r01.scale+tlrc.BRIK
end
3dTstat -prefix rm.mean_r02 pb03.sub_08.r02.blur+tlrc
++ 3dTstat: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: KR Hammett & RW Cox
++ Output dataset ./rm.mean_r02+tlrc.BRIK
3dcalc -a pb03.sub_08.r02.blur+tlrc -b rm.mean_r02+tlrc -c mask_epi_extents+tlrc -expr c * min(200, a/b*100)*step(a)*step(b) -prefix pb04.sub_08.r02.scale
++ 3dcalc: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ Authored by: A cast of thousands
++ Output dataset ./pb04.sub_08.r02.scale+tlrc.BRIK
end
gen_epi_review.py -script @epi_review.sub_08 -dsets pb00.sub_08.r01.tcat+orig.HEAD pb00.sub_08.r02.tcat+orig.HEAD
rm -f rm.epi.1.r01+tlrc.BRIK rm.epi.1.r01+tlrc.HEAD rm.epi.1.r02+tlrc.BRIK rm.epi.1.r02+tlrc.HEAD rm.epi.all1+orig.BRIK rm.epi.all1+orig.HEAD rm.epi.mean+tlrc.BRIK rm.epi.mean+tlrc.HEAD rm.epi.min.r01+tlrc.BRIK rm.epi.min.r01+tlrc.HEAD rm.epi.min.r02+tlrc.BRIK rm.epi.min.r02+tlrc.HEAD rm.epi.nomask.r01+tlrc.BRIK rm.epi.nomask.r01+tlrc.HEAD rm.epi.nomask.r02+tlrc.BRIK rm.epi.nomask.r02+tlrc.HEAD rm.epi.volreg.r01+orig.BRIK rm.epi.volreg.r01+orig.HEAD rm.epi.volreg.r02+orig.BRIK rm.epi.volreg.r02+orig.HEAD rm.mask_r01+tlrc.BRIK rm.mask_r01+tlrc.HEAD rm.mask_r02+tlrc.BRIK rm.mask_r02+tlrc.HEAD rm.mean_r01+tlrc.BRIK rm.mean_r01+tlrc.HEAD rm.mean_r02+tlrc.BRIK rm.mean_r02+tlrc.HEAD rm.resam.anat+tlrc.BRIK rm.resam.anat+tlrc.HEAD rm.resam.group+tlrc.BRIK rm.resam.group+tlrc.HEAD
if ( -e @ss_review_basic ) then
cd ..
echo execution finished: `date`
date
execution finished: Tue Nov 4 03:39:43 UTC 2025
While the preprocessing is running, here’s a brief overview of the key steps. For a more detailed explanation of each preprocessing stage, have another look at Andy’s Brain Book’s excellent tutorial.
Slice Time Correction#
Even though slice timing correction is often applied to functional data, in this dataset the slices were already aligned in time.
When running 3dTshift, AFNI detected this and simply created a copy of the input data without modification (as indicated by the warning message).
Normally, if slice timing correction were needed, it would use the first slice as the reference (specified by the -tzero 0 option in 3dTshift).
You can still see the slice timing correction step in the proc.sub_08 script, but in this case, it had no effect on the data.
Note: Although slice-timing correction is common, there are some general objections: it involves interpolating the data (which is best avoided when possible), it may not significantly improve results when the TR is short (e.g., around 1 second), and timing differences can sometimes be modeled later using a temporal derivative during model fitting.
# =================== tshift ===================
foreach run ( $runs )
3dTshift -tzero 0 -quintic -prefix pb01.$subj.r$run.tshift \
pb00.$subj.r$run.tcat+orig
end
Registration and Normalization#
Registration aligns the functional and anatomical images. It usually begins by assuming the images are roughly in the same space; if not, their outlines are aligned. To fine-tune the alignment, the algorithm uses differences in contrast (mutual information), matching dark areas on one image with bright areas on the other. AFNI’s preferred method is Local Pearson Correlation (LPC), which gives greater weight to brighter areas in the functional data. Once a good alignment is found, the same transformations can later be applied to warp the functional images to a standard template.
In this example, AFNI’s align_epi_anat.py command is used to perform the registration. Although this tool can also handle slice-timing correction and functional volume alignment, it will be used here only for registration
The key options used are:
-anat2epi: Aligns the anatomical image to the functional image (not the other way around), minimizing changes to the functional data.-suffix_al_junk: Adds a suffix to intermediate files, which will be useful for later steps.-epi,-epi_base,-epi_strip: Chooses the functional volume with least variability as the reference and strips non-brain tissue using 3dAutomask.-giant_move: Helps find an initial rough alignment if the images are very misaligned.-volreg off,-tshift off: Indicates that alignment and slice-timing correction are not included in the current command.
Registration with align_epi_anat.py#
=================== align ====================
align_epi_anat.py -anat2epi -anat sub-08_T1w+orig \
-save_skullstrip -suffix _al_junk \
-epi vr_base_min_outlier+orig -epi_base 0 \
-epi_strip 3dAutomask \
-giant_move \
-volreg off -tshift off
Normalization with @auto_tlrc#
After registration, the anatomical image is normalized to a standard template (MNI_avg152T1) using AFNI’s @auto_tlrc command. Since the anatomical image has already been skull-stripped, the -no_ss option is used. Because the centers of the anatomical and template images were far apart, the -init_xform AUTO_CENTER option was included.
This helps by roughly aligning the centers of the two images before the fine registration step.
The transformation needed to align the anatomical image to the template is stored as an affine matrix in the header of the anatomical iamge.
This matrix is then extracted with the cat_matvec command and saved as warp.anat.Xat.1D, so it can later be applied to the functional images as well.
#warp anatomy to standard space @auto_tlrc -base MNI_avg152T1+tlrc -input sub-08_T1w_ns+orig -no_ss -init_xform AUTO_CENTER#store forward transformation matrix in a text file cat_matvec sub-08_T1w_ns+tlrc::WARP_DATA -I > warp.anat.Xat.1D
Alignment and Motion Correction#
Motion correction is done with AFNI’s 3dvolreg command.
The functional volumes are aligned to a reference image, which is selected as the volume with the fewest outliers (identified using 3dToutcount earlier).
Motion parameters are saved into a text file (-1Dfile) and the corresponding affine transformation matrices into another file (-1Dmatrix_save).
# =================== volreg ===================
#register and warp
foreach run ( $runs )
3dvolreg -verbose -zpad 1 -base vr_base_min_outlier+orig \
-1Dfile dfile.r$run.1D -prefix rm.epi.volreg.r$run \
-cubic \
-1Dmatrix_save mat.r$run.vr.aff12.1D \
pb01.$subj.r$run.tshift+orig
These motion correction matrices are then concatenated with the matrices from anatomical registration and normalization.
#catenate volreg/epi2anat/tlrc xforms
cat_matvec -ONELINE \
{$subj}_T1w_ns+tlrc::WARP_DATA -I \
{$subj}_T1w_al_junk_mat.aff12.1D -I \
mat.r$run.vr.aff12.1D > mat.r$run.warp.aff12.1D
Using 3dAllineate and the -1Dmatrix_apply option, both motion correction and normalization are applied to the functional images in one step.
#warp the all-1 dataset for extents masking
3dAllineate -base {$subj}_T1w_ns+tlrc \
-input rm.epi.all1+orig \
-1Dmatrix_apply mat.r$run.warp.aff12.1D \
-mast_dxyz 3 -final NN -quiet \
-prefix rm.epi.1.r$run
Smoothing#
Smoothing is done with AFNI’s 3dmerge command, which can be found under the “blur” header.
The -1blur_fwhm option applies spatial smoothing with a 4mm kernel, and -doall ensures smoothing is done across all volumes. After smoothing, the images are scaled to a mean intensity of 100, allowing changes to be interpreted as percent signal change. Finally, a brain mask is applied to remove non-brain voxels, preparing the data for statistical analysis.
# ==================== blur ====================
#blur each volume of each run
foreach run ( $runs )
3dmerge -1blur_fwhm 4.0 -doall -prefix pb03.$subj.r$run.blur \
pb02.$subj.r$run.volreg+tlrc
end
Masking and Scaling#
Masking#
fMRI datasets include not only brain voxels but also irrelevant areas like the skull, neck, and air. To focus analysis and speed up processing, a mask is applied: voxels inside the mask are kept, and those outside are discarded. AFNI’s 3dAutomask creates these masks by simply specifying the input and output datasets.
# ==================== mask ====================
# create 'full_mask' dataset (union mask)
foreach run ( $runs )
3dAutomask -prefix rm.mask_r$run pb03.$subj.r$run.blur+tlrc
end
The additional steps within the “mask” block combine individual fMRI masks into a union mask, compute a mask for the anatomical image, and take the intersection of both to define the final analysis region.
Scaling#
fMRI signal intensities are arbitrary and vary between subjects and runs. To make meaningful comparisons, AFNI scales each voxel’s time series to a mean of 100, allowing for consistent signal intensity contrasts between conditions and across subjects.
# ==================== scale ====================
# scale each voxel time series to have a mean of 100
# (be sure no negatives creep in)
# (subject to a range of [0,200])
foreach run ( $runs )
3dTstat -prefix rm.mean_r$run pb03.$subj.r$run.blur+tlrc
3dcalc -a pb03.$subj.r$run.blur+tlrc -b rm.mean_r$run+tlrc \
-c mask_epi_extents+tlrc \
-expr 'c * min(200, a/b*100)*step(a)*step(b)' \
-prefix pb04.$subj.r$run.scale
end
Effect of Scaling: Before scaling, time-series values are arbitrary and vary between subjects. After scaling, each subject’s data is normalized to the same mean, enabling consistent comparisons across runs and subjects. To visualize both the original and scaled data with Matplotlib, the AFNI format needs to be converted to NIfTI format using the 3dAFNItonNIFI command.
The following plot compares the signal intensity time series from a voxel at the center of the brain, before and after scaling. The red dashed line indicates the mean signal intensity for each time series.
!3dAFNItoNIFTI -prefix ./afni_processing/sub_08.results/pb03.sub_08.r01.blur.nii.gz ./afni_processing/sub_08.results/pb03.sub_08.r01.blur+tlrc
!3dAFNItoNIFTI -prefix ./afni_processing/sub_08.results/pb04.sub_08.r01.scale.nii.gz ./afni_processing/sub_08.results/pb04.sub_08.r01.scale+tlrc
++ 3dAFNItoNIFTI: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: varying brick factors, writing NIfTI as float
++ 3dAFNItoNIFTI: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: varying brick factors, writing NIfTI as float
blur_nii = nib.load("./afni_processing/sub_08.results/pb03.sub_08.r01.blur.nii.gz").get_fdata()
scale_nii = nib.load("./afni_processing/sub_08.results/pb04.sub_08.r01.scale.nii.gz").get_fdata()
# Choose a voxel (e.g., center)
x, y, z = blur_nii.shape[0]//2, blur_nii.shape[1]//2, blur_nii.shape[2]//2
time_series1 = blur_nii[x, y, z, :]
time_series2 = scale_nii[x, y, z, :]
n_timepoints = time_series1.shape[0]
# Calculate the means of the time series
mean_time_series1 = time_series1.mean()
mean_time_series2 = time_series2.mean()
# Create subplots
fig, axs = plt.subplots(2, 1, figsize=(10, 6), sharex=True)
axs[0].plot(time_series1, color='blue')
axs[0].set_title("Signal intensity before scaling")
axs[0].set_ylabel("Signal Intensity")
axs[0].axhline(mean_time_series1, color='red', linestyle='--', linewidth=0.7, label=f"Mean: {mean_time_series1:.2f}")
axs[0].grid(True)
axs[0].legend()
axs[0].set_xlim(0, n_timepoints - 1)
axs[1].plot(time_series2, color='orange')
axs[1].set_title("Signal intensity after scaling")
axs[1].set_xlabel("Timepoint")
axs[1].set_ylabel("Signal Intensity")
axs[1].axhline(mean_time_series2, color='red', linestyle='--', linewidth=0.7, label=f"Mean: {mean_time_series2:.2f}")
axs[1].grid(True)
axs[1].legend()
axs[0].set_xlim(0, n_timepoints - 1)
plt.tight_layout()
plt.show()
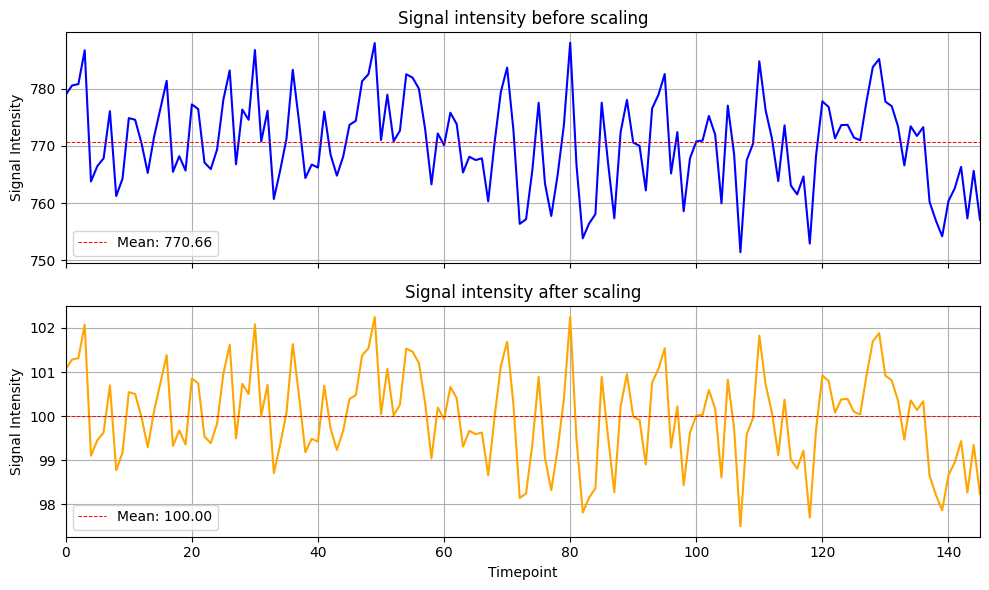
Results: Checking Preprocessing#
Let’s have a look at the results directory. The files containing the pb string are the preprocessed functional images at each preprocessing step, and the files with the T1w string are the preprocessed anatomical images. Auxiliary functional images are created to assist with specific preprocessing steps, and auxiliary text files contain information about transformation matrices and movement parameter:
!ls ./afni_processing/sub_08.results
@epi_review.sub_08 pb01.sub_08.r01.tshift+orig.HEAD
anat_final.sub_08+tlrc.BRIK pb01.sub_08.r02.tshift+orig.BRIK
anat_final.sub_08+tlrc.HEAD pb01.sub_08.r02.tshift+orig.HEAD
anat_w_skull_warped+tlrc.BRIK pb02.sub_08.r01.volreg+tlrc.BRIK
anat_w_skull_warped+tlrc.HEAD pb02.sub_08.r01.volreg+tlrc.HEAD
dfile.r01.1D pb02.sub_08.r02.volreg+tlrc.BRIK
dfile.r02.1D pb02.sub_08.r02.volreg+tlrc.HEAD
dfile_rall.1D pb03.sub_08.r01.blur+tlrc.BRIK
final_epi_vr_base_min_outlier+tlrc.BRIK pb03.sub_08.r01.blur+tlrc.HEAD
final_epi_vr_base_min_outlier+tlrc.HEAD pb03.sub_08.r01.blur.nii.gz
full_mask.sub_08+tlrc.BRIK pb03.sub_08.r02.blur+tlrc.BRIK
full_mask.sub_08+tlrc.HEAD pb03.sub_08.r02.blur+tlrc.HEAD
mask_anat.sub_08+tlrc.BRIK pb04.sub_08.r01.scale+tlrc.BRIK
mask_anat.sub_08+tlrc.HEAD pb04.sub_08.r01.scale+tlrc.HEAD
mask_epi_anat.sub_08+tlrc.BRIK pb04.sub_08.r01.scale.nii.gz
mask_epi_anat.sub_08+tlrc.HEAD pb04.sub_08.r02.scale+tlrc.BRIK
mask_epi_extents+tlrc.BRIK pb04.sub_08.r02.scale+tlrc.HEAD
mask_epi_extents+tlrc.HEAD pre.sub-08_T1w_ns+orig.BRIK
mask_group+tlrc.BRIK pre.sub-08_T1w_ns+orig.HEAD
mask_group+tlrc.HEAD pre.sub-08_T1w_ns_WarpDrive.log
mat.basewarp.aff12.1D stimuli
mat.r01.vr.aff12.1D sub-08_T1w+orig.BRIK
mat.r01.warp.aff12.1D sub-08_T1w+orig.HEAD
mat.r02.vr.aff12.1D sub-08_T1w_al_junk+orig.BRIK
mat.r02.warp.aff12.1D sub-08_T1w_al_junk+orig.HEAD
motion_sub_08_enorm.1D sub-08_T1w_al_junk_mat.aff12.1D
out.allcostX.txt sub-08_T1w_ns+orig.BRIK
out.mask_ae_dice.txt sub-08_T1w_ns+orig.HEAD
out.mask_ae_overlap.txt sub-08_T1w_ns+tlrc.BRIK
out.mask_at_dice.txt sub-08_T1w_ns+tlrc.HEAD
out.min_outlier.txt sub-08_T1w_ns.Xaff12.1D
out.pre_ss_warn.txt sub-08_T1w_ns.Xat.1D
outcount.r01.1D sub-08_T1w_ns.maskwarp.Xat.1D
outcount.r02.1D sub-08_T1w_ns_shft.1D
outcount_rall.1D volumized+tlrc.BRIK
pb00.sub_08.r01.tcat+orig.BRIK volumized+tlrc.HEAD
pb00.sub_08.r01.tcat+orig.HEAD vr_base_min_outlier+orig.BRIK
pb00.sub_08.r02.tcat+orig.BRIK vr_base_min_outlier+orig.HEAD
pb00.sub_08.r02.tcat+orig.HEAD warp.anat.Xat.1D
pb01.sub_08.r01.tshift+orig.BRIK
In AFNI, a +tlrc extension (and the “Talairach View”) indicates that the image has been normalized. However, this does not necessarily mean the image is in Talairach space. The Talairach label is retained for legacy reasons, ensuring compatibility with older versions of the software. To verify which space the image has been warped to, you can use the 3dinfo command and check the “Template Space” field. The three possibilities are: “ORIG” (the image has not been warped), “TLRC” (normalized to Talairach space), and “MNI” (normalized to MNI space).
Visualizations#
To visualize the preprocessing outputs with ipyniivue, the files are first converted to NIfTI format. Once loaded, check whether the functional and anatomical images are properly aligned to MNI space. Misalignments could indicate issues with the preprocessing steps. Smoothed images will generally appear less detailed than the original images due to the blurring effect; this is expected, but they should not be excessively blurry or too sharp. The scaled functional image will have less anatomical definition, as the signal intensity has been normalized across time, making it more uniform across brain voxels. When reviewing the masks, ensure they appropriately cover the brain regions of interest and match the underlying anatomy. For example, the “mask_group” is typically a more liberal mask compared to the “full_mask”.
!3dAFNItoNIFTI -prefix ./afni_processing/mni152.nii.gz /cvmfs/neurodesk.ardc.edu.au/containers/afni_21.2.00_20210714/afni_21.2.00_20210714.simg/opt/afni-latest/MNI_avg152T1+tlrc
!3dAFNItoNIFTI -prefix ./afni_processing/sub_08.results/anat_w_skull_warped.nii.gz ./afni_processing/sub_08.results/anat_w_skull_warped+tlrc
!3dAFNItoNIFTI -prefix ./afni_processing/sub_08.results/anat_final.nii.gz ./afni_processing/sub_08.results/anat_final.sub_08+tlrc
!3dAFNItoNIFTI -prefix ./afni_processing/sub_08.results/pb02.sub_08.r01.volreg+tlrc.nii.gz ./afni_processing/sub_08.results/pb02.sub_08.r01.volreg+tlrc
!3dAFNItoNIFTI -prefix ./afni_processing/sub_08.results/full_mask.sub_08+tlrc.nii.gz ./afni_processing/sub_08.results/full_mask.sub_08+tlrc
!3dAFNItoNIFTI -prefix ./afni_processing/sub_08.results/mask_group.nii.gz ./afni_processing/sub_08.results/mask_group+tlrc
++ 3dAFNItoNIFTI: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dAFNItoNIFTI: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dAFNItoNIFTI: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dAFNItoNIFTI: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
*+ WARNING: varying brick factors, writing NIfTI as float
++ 3dAFNItoNIFTI: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
++ 3dAFNItoNIFTI: AFNI version=AFNI_21.2.00 (Jul 8 2021) [64-bit]
#extract the first volume
index_img('./afni_processing/sub_08.results/pb02.sub_08.r01.volreg+tlrc.nii.gz', 0).to_filename('./afni_processing/sub_08.results/pb02.sub_08.r01.volreg+tlrc_vol0.nii.gz')
# Paths to files
anat_skull = './afni_processing/sub_08.results/anat_w_skull_warped.nii.gz'
anat_final = './afni_processing/sub_08.results/anat_final.nii.gz'
volreg_func = './afni_processing/sub_08.results/pb02.sub_08.r01.volreg+tlrc_vol0.nii.gz'
mni_nifti = './afni_processing/mni152.nii.gz'
full_mask = './afni_processing/sub_08.results/full_mask.sub_08+tlrc.nii.gz'
group_mask = './afni_processing/sub_08.results/mask_group.nii.gz'
nv = NiiVue()
# Paths to all volumes
volumes = [
{"path": mni_nifti, "colormap": "gray", "opacity": 1.0}, # MNI background
{"path": anat_final, "colormap": "red", "opacity": 0.7}, # default overlay
{"path": volreg_func, "colormap": "red", "opacity": 0.0},
{"path": full_mask, "colormap": "green", "opacity": 0.0},
{"path": group_mask, "colormap": "blue", "opacity": 0.0},
]
# Load all volumes into viewer
nv.load_volumes(volumes)
# Dropdown options
overlay_dropdown = widgets.Dropdown(
options=[
("Anatomical aligned to MNI", 1),
("Motion-corrected functional to MNI", 2),
("Union of functional masks (full_mask)", 3),
("Dilated group mask (group_mask)", 4)
],
value=1,
description='Overlay:',
style={'description_width': 'initial'},
layout=widgets.Layout(width='350px')
)
# Update function
def update_overlay(change):
selected_idx = change.new
# Set all overlays to 0 opacity
for idx in range(1, len(volumes)):
nv.volumes[idx].opacity = 0.0
# Activate the selected one
nv.volumes[selected_idx].opacity = 0.7
# Attach the observer
overlay_dropdown.observe(update_overlay, names='value')
# Show viewer and dropdown
display(VBox([overlay_dropdown, nv]))
Image(url='https://raw.githubusercontent.com/NeuroDesk/example-notebooks/refs/heads/main/books/images/afni_preproc_alignment.png')
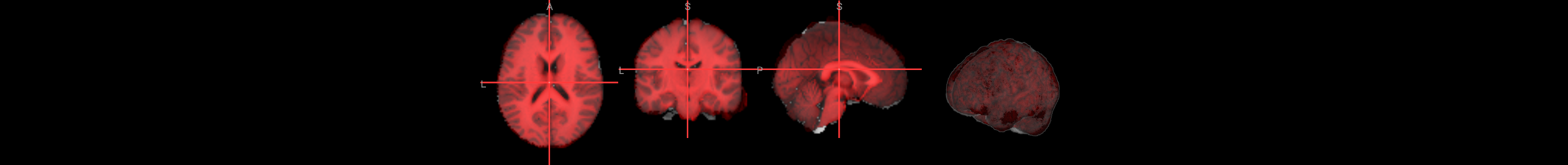
Dependencies in Jupyter/Python#
Using the package watermark to document system environment and software versions used in this notebook
%load_ext watermark
%watermark
%watermark --iversions
Last updated: 2025-11-04T03:40:18.797246+00:00
Python implementation: CPython
Python version : 3.11.6
IPython version : 8.16.1
Compiler : GCC 12.3.0
OS : Linux
Release : 5.4.0-204-generic
Machine : x86_64
Processor : x86_64
CPU cores : 32
Architecture: 64bit
ipywidgets: 8.1.2
nibabel : 5.3.2
numpy : 2.2.4
ipyniivue : 2.3.2
IPython : 8.16.1
matplotlib: 3.8.4
nilearn : 0.11.1
